Analysis of KRAS Gene Mutations by using of Circulating Tumor DNA
Author'(s): Satoshi Tokuda1, Hajime Orita1*, Shunsuke Sakuraba1, Tomoaki Ito1 , Hideo Shimizu1, Mutsumi Sakurada1, Tomoyuki Kushida1, Kenichiro Tanaka1, Hiroshi Maekawa1, Ryo Matoba2 and Koichi Sato1
1 Department of Surgery, Juntendo Shizuoka Hospital, Shizuoka,Japan.
2 DNA chip Research Inc., Shizuoka, Japan.
*Correspondence:
Hajime Orita, MD, PhD, Department of Surgery, Juntendo Shizuoka Hospital, Shizuoka, Japan, Tel: 055-948-3111; Fax: 055-946-0514; E-mail: oriori@juntendo.ac.jp.
Received: 18 April 2018; Accepted: 21 May 2018
Citation: Satoshi Tokuda, Hajime Orita, Shunsuke Sakuraba, et al. Analysis of KRAS Gene Mutations by using of Circulating Tumor DNA. Cancer Sci Res. 2018; 1(2): 1-4.
Abstract
Introduction: The treatment for advanced colorectal cancer (CRC) is getting better owing to the progression of molecular target drugs and one of the targets is Epidermal Growth Factor Receptor (EGFR). To detect the gene mutation, we used circulating tumor DNA (ctDNA) in the blood to confirm whether we can identify KRAS mutations from serums.
Materials and Methods: We gathered serum samples from 20 patients (2013-2015) who had undergone colon cancer operations. We abstracted ctDNA from the serum samples, examined the existence of KRAS mutations, and then compared them to the biopsy specimen.
Results and Discussion: 8 cases are wild type and 7 cases are mutation type for both (biopsy specimen and serum), however 5 cases are mutation type for biopsy specimen and wild type for serum. We show here that the sensitivity of this study for testing KRAS mutations is 83.3%.
Conclusion: This study suggests that the possibility of using the ctDNA in serum samples to check for the KRAS mutation.
Keywords
Introduction
The treatment for cancer is getting better owing to the discovery of new biomarkers; for example epidermal growth factor receptor (EGFR), kirsten rat sarcoma viral oncogene homolog (KRAS) gene, and the echinoderm microtubule-associated protein like 4–anaplastic lymphoma receptor tyrosine kinase (EML4–ALK) fusion gene. Colorectal cancer (CRC) is the third most number of cancer and tumor-related death [1]. Not only surgical resection but also chemotherapy is important for the treatment of CRC and the EGFR is one of the therapeutic targets. That is to say, we can use the anti-EGFR monoclonal antibodies in patients with RAS wild type tumors [2,3], so we have to check gene mutations before using anti-EGFR monoclonal antibodies. Even if the primary focus does not have RAS mutations, it is not the end of the matter. There are various cases, for example, in which the metastatic focus has the mutation and the cancer gets the mutation under treatment, thus frequent monitoring is required. One of the reasons is the intratumoral heterogeneity and many cancers will get resistance as a result of the heterogeneity [4]. In addition, we can only obtain a little information from tumor tissue and sampling is sometimes difficult.
Some papers revealed the usefulness of liquid biopsy to detect the gene mutations [5-7], however we have yet to reach a consensus.
In this study, we used circulating tumor DNA (ctDNA) in the blood, confirmed whether we can identify KRAS mutations from serums and made sure of the sensitivity. Incidentally, we started this study before NRAS and BRAF came into the spotlight, thus focusing solely on KRAS mutations.
Liquid biopsy
Some paper showed the usability of liquid biopsy. The report of using BEAMing (beads, emulsions, amplification, and magnetics), the prevalence of RAS mutations detected in plasma (51%) vs. tumor (53%), RAS analysis revealed high concordance of plasma and tissue results of 91.8% [8]. Besides, it is said that during the CRC treatment with anti-EGFR antibodies, the CRC gene acquires the resistance [9-12].
By using of liquid biopsy, J Trojan et al. could detect the acquired KRAS mutation early and change the treatment plan [13].
Methods
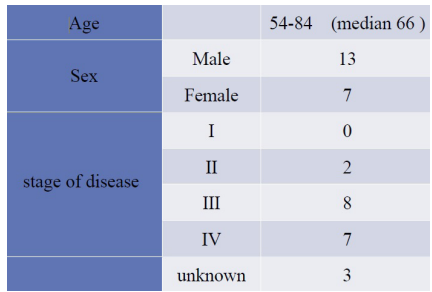
We gathered serum samples from 20 patients (2013-2015) who had undergone colon cancer operations. All patients were Japanese and their stages were ~. We abstracted ctDNA from the serum samples, examined the existence of KRAS mutations, and then compared them to primary and metastatic focus (biopsy specimen). The biopsy was routine diagnostic analysis. Clinical and pathological features of patients are shown in Table 2. We collected peripheral blood from the patients before their operation and the blood was stored at -80.
We utilized the next generation sequencing (NGS) to check for KRAS mutations.
Extraction of the cfDNA from the serum samples
We centrifuged the serum samples at 800×g for 10 minutes at room temperature. After pouring the supernatant in a new tube, we centrifuged it at 15100×g for 10 minutes at room temperature. After pouring the supernatant in a new tube again, cfDNA was extracted from it with the Qiagen Circulating Nucleic Acid kit (QIAGEN, Germany). The cfDNA was measured with TapeStation HighSensitivity D1000kit (Agilent Technologies, Inc, USA).
Library Preparation
The amplification of KRAS gene was performed by using of oligo DNA primer. Library sequencing was prepared by 2 Step Multiplex PCR.
1st round PCR reactions were run on a 96-well plate with adaptor sequence of Illumina Miseq system, PCR Primer that is KRAS gene specific sequence (each 5uM; Fasmac Co., Ltd. Japan), PCR reagent (KOD -Plus- 200U (TOYOBO, Japan); 10x KOD buffer, 2mM dNTP, 25mM MgSO4, KOD -Plus- (1.0 U/μl) ) and cfDNA sample (10μl); we sealed the liquid surface with mineral oil.
We used the Veriti™ Dx Thermal Cycler (Thermo Fisher Scientific, USA) under the following condition: 94 2 min, (94 15 sec, 55 30 sec, 68 15 sec) 20Cycles, 10 hold. 2ndround PCR was performed as bellow: we added the 2nd PCR Primer (1st round PCR primer+Index+sequence primer) to the plate after the 1st round PCR so as not to disturb the oil surface. After that we did PCR again. The reaction condition was 94 2minutes, (94 15 seconds, 55 30 seconds, 68 15 seconds) 20 cycles, 10 hold.
PCR Product was refined with Mini Elute PCR Purification Kit (QIAGEN, Germany).
For the sequence analysis, we used Miseq System (Illumina ,USA; Pair end run) with Reagent Cartridge v2. To confirm the detail of the operating procedure, we referred to Miseq System Quick Ref Guide.
All patients of this study signed written informed consent forms.
Results
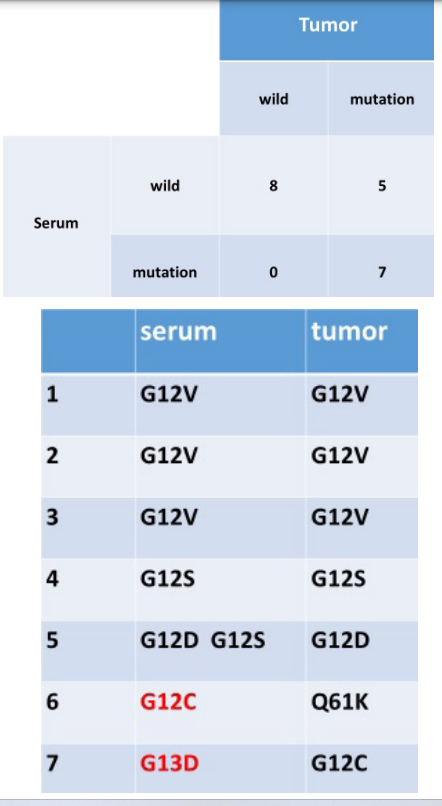
We retrospectively studied 20 patients who had undergone surgery of CRC during 2013-2015. Their stages are -. We show the result of this study as below (Table 1). The summary of the result is Figure 1. 8 cases are wild type and 7 cases are mutation type for both (biopsy specimen and serum), however 5 cases are mutation type for biopsy specimen and wild type for serum. We focused on the 7 cases of mutation type for both and compared the mutation (Figure 2). Patient 1 to 5 (patient 5 has two mutations in serum) are the same mutation. Conversely, patient 6 and 7 differ between biopsy specimen and serum.

Next we considered about the 5 cases; differ from each other. The reason is the problem of the quality of the specimens. 4 out of the 5 were hemolytic specimens. In the 4 cases the mixed normal genome might have hindered the sequence of the KRAS mutation in the serum. We excluded the poor specimens (8 out of the 20) and recounted (Figure 3). Figure 3 shows high concordance rate between biopsy specimen and serum. We show here that the sensitivity of this study for testing KRAS mutations is 83.3%.

Discussion
To detect CRC, we utilize various approaches; serum markers, Computed Tomography (CT) scans and Magnetic Resonance Imaging (MRI). Add to that, we may be able to use the ctDNA in the near future. CtDNA in the blood consists of DNA derived from cancer tissues. CtDNA is included in cell free DNA in the blood. Average size is about 166 bp, half-life is about 1 hour for the rapid phase and a second phase of 13 hours. This approach which uses the ctDNA is called ‘liquid biopsy’. It is utilized in various situations as stated above. Many benefits can be gained from ‘liquid biopsy’. First, the liquid biopsy is useful for the diagnosis of cancer; for example lung cancer [1]. The biopsy of the lung cancer is sometimes difficult compared to intestinal cancer. Similarly, we can utilize this technique for the diagnosis of pancreatic solid masses [3].
Second, we can monitor the recurrence or metastasis of CRC after surgery. According to the study of Reinert et al., they could detect incipient recurrence of CRC by ctDNA. Their approach provided 2-15 (mean 10) months' lead time on detection of metastatic recurrence compared to conventional follow-up [14].
In this study, we used ctDNA and confirmed whether KRAS mutations can be identified from serums. The sensitivity is 83.3%, which a fair is resulting although one that does raise some concerns. We focused on Figure 2 and considered patients 6 and 7. Why was the mutation area mismatched in these 2 cases? The reason for patient 6 may have been due to our lack of a measurement method. We had no method check the mutation of Q61K in serum. Regarding patient 7, we assumed the involvement of the tumor heterogeneity. According to the study of Vivian et al., tumor heterogeneity is revealed by different KRAS mutations in the tumor center versus periphery [15].
There is the difference of gene mutation between surface and internal. It indicates the difficulty of the treatment of CRC. As for patient 5, this patient has 2 KRAS mutations in serum. We think that there are 2 possibilities. 1 possibility is that the patient had another carcinoma besides the CRC. However he had no another carcinoma in preoperative examination, therefore the hypothesis is low possibility. The second possibility is the tumor heterogeneity. Vivian et al. mentioned the presence of two different KRAS mutations within the same tumor. Several limitations of this study should be acknowledged.
First, the quality of the specimens was a big problem and we could not detect the KRAS mutations from the poor serum samples. 8 out of 20 were hemolytic specimens and 3 of them were Stage patients. The previous study of the NSCLC patients with extra- thoracic metastatic disease (M1b) showed high sensitivity of the NGS-assay compared to M1a and M0 [1]. Even the Stage , we were not able to detect the KRAS mutation with hemolytic specimens.
Second, the limitations of this study include small sample size. We believe that an increase in number of the subjects would make our conclusion sounder.
Conclusions
This study suggests that it is possible to use ctDNA in serum samples to check for the KRAS mutation. The first and second line therapies for unresectable CRC are consisted of a fluoropyrimidine doublet (FOLFOX or FOLFIRI) combined with the anti-EGFR monoclonal antibodies or the anti-vascular endothelial growth factor monoclonal antibodies [2,3]. In that case, the detection of KRAS mutation will be a big problem. Using the technique of liquid biopsy, we can treat cancer patients with more precision.
References
- Siegel RL, Miller KD, Jemal Cancer Statistics. 2016; 66: 7-30.
- Van Cutsem E, Cervantes A, Adam R, et ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. 2016; 27: 1386-1422.
- Al B. Benson, Alan P. Venook, Lynette Cederquist, et al. Colon Cancer. Clinical Practice Guidelines in Oncology. 2017; 15: 370-398.
- Heitzer E, Ulz P, Geigl Reviews Circulating Tumor DNA as a Liquid Biopsy for Cancer. 2015; 61: 112-123.
- Rachiglio AM, Esposito Abate R, Sacco A, et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. 2016; 7: 66595-
- Malignancies NIH Public Access. 2014; 6: 224.
- Kinugasa H, Nouso K, Miyahara K, et al. Detection of K-ras Gene Mutation by Liquid Biopsy in Patients With Pancreatic Cancer. 2015; 121: 2271-2280.
- Schmiegel W, Scott RJ, Dooley S, et Blood-based detection of RAS mutations to guide anti- EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol. 2017; 11: 208-219.
- Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and Chemotherapy as Initial Treatment for Metastatic Colorectal Cancer. N Engl J Med. 2009; 360: 1408-1417.
- Tu D, Tebbutt NC, Simes RJ, et al. New England Journal. 2008; 1757-1765.
- Douillard JY, Oliner KS, Siena S, Tabernero J, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013; 369: 1023-1034.
- Diaz LA, Jr, Williams RT, Wu J, et The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012; 486: 537-540.
- Trojan J, Klein-scory S, Koch C, et Case Report Metastatic Colorectal Cancer. 2017; 1-3.
- Reinert T, Schøler LV, Thomsen R, et Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016; 5: 625-634.
- Mutation H, Kosmidou V, Hellenic N, et al. Tumor heterogeneity revealed by KRAS, BRAF, and PIK3CA pyrosequencing: KRAS and PIK3CA intratumor mutation profile differences and their therapeutic implications. Hum Mutat. 2014; 35: 329-340.