Characterization of the Interstitial Cystitis/Bladder Pain Syndrome Microbiome in Clinically Diagnosed Patients
Author'(s): Alexander W. Larsen1, Yuan Chen1, Keith A. Crandall1,2, Crystal R. Icenhour1 and C. Alexander Valencia1,3*
1 Aperiomics, Sterling, Virginia, US.
2 Computational Biology Institute, Milken Institute School of Public Health, George Washington University, Washington, DC 20052, US.
3Interpath Laboratory, Pendleton, Oregon, US.
*Correspondence:
Dr. C Alexander Valencia, Address: 2460 SW Perkins Ave. Pendleton, OR 97801, US.
Received: 06 Jul 2022; Accepted: 20 Jul 2022; Published: 25 Jul 2022
Citation: Larsen NW, Chen Y, Crandall KA, et al. Characterization of the Interstitial Cystitis/Bladder Pain Syndrome Microbiome in Clinically Diagnosed Patients. Clin Immunol Res. 2022; 6(1): 1-9.
Abstract
Aim: Ongoing research in the urinary disease field suggests that a subset of patients with bladder pain and incontinence may have an infection as the cause of those clinical features, which may be difficult to diagnose by standard testing. Shotgun metagenomics sequencing (smNGS) provides a comprehensive way to identify infectious agents, namely, viruses, fungi, or bacteria, one possible cause that may be responsible for Interstitial Cystitis/Bladder Pain Syndrome (IC/BPS).
Methods: DNA was extracted from these samples and sequenced by combining KAPA HyperPlus library construction with next generation sequencing (Nextseq500, Illumina). Reads were analyzed using Xplore-Patho®, an analytical system that permits the detection of 37,000+ microorganisms, including over 12,000 known pathogens.
Results: Urine samples were collected from 49 clinically diagnosed IC/BPS. Additionally, controls urine samples were collected from 13 individuals without chronic lower urinary tract symptoms. Comparisons in diversity between the IC/ BPS and control groups were performed. Of the 41 female patients who were diagnosed with IC/BPS, 35 had signs of infection with a microbial signature for IC/BPS, including the presence of Proteobacterium, BK and JC viruses (Betapolyomavirus), and higher fungal loads. The alpha diversity differences between the female IC/BPS and female control groups were not statistically significant (p = 0.18). In contrast, the beta diversity was found to be different between female IC/BPS and control groups using Kruskal-Wallis ANOVA (p = 2.33e-14).
Conclusion: Culture, PCR, antigen, or antibody-based standard urine tests cannot comprehensively screen for the infectious causes of IC/BPS. We present an all-inclusive, unbiased smNGS approach that helped unravel the infectious causes of IC/BPS. This new tool can help clinicians treat their patients in a more timely and effective manner.
Keywords
Introduction
Interstitial cystitis/bladder pain syndrome (IC/BPS) is a pain disorder with that exhibits urine storage problems. The International Continence Society (ICS) and the European Society for the Study of IC/BPS (ESSIC) define this condition as having chronic (>6 months) pelvic pain associated with urinary bladder in conjunction with at least one other urinary symptom [1]. Urinary tract infection (UTI), overactive bladder, hypersensitive bladder, chronic urethral syndrome, vulvodynia, prostatitis and endometriosis are difficult to treat because they have overlapping symptomology. The IC/ BPS population includes approximately 3 to 8 million women and 1 to 4 million men in the United States [2].
The bladder-specific microbiome has been defined and the abundance and species richness has been hypothesized to be involved in overactive bladder syndrome (OAB) and IC/BPS symptomology [3,4]. In addition, there has been a question of traditional colony forming unit thresholds for UTI confirmation, and association between UTI and OAB was explored [5]. Patients with OAB may have possessed urogenital infections that were missed by mid-stream urine cultures [6]. Indeed, a significant number of patients with refractory idiopathic detrusor overactivity had bacteriuria in contrast to controls [7]. Moreover, OAB patients typically had higher infected urothelial cell counts and microscopic pyuria compared to controls [8]. A uropathogenic E. coli infection releases cytokines and interleukins that initiate an immune response, immune cell infiltration and enhance bladder sensation [9,10]. Recently, a study demonstrated that combination antibiotic treatment improved OAB symptoms [11]. A changes in bacterial species of the bladder microbiome were associated with OAB and IC/PBS [12]. Specifically, women with IC/PBS, but not OAB, had a less diverse microbiome [12-14]. Similarly, a decrease in Lactobacillus spp. was noted in OAB and IC/BPS patients, the absence of Lactobacillus acidophilus was associated with higher pain scores and Proteus was detected in patients with OAB and urinary tract symptoms [12,15]. The data demonstrated an interaction between the urinary system and afferent nerves potentially mediated by the urothelium.
Shotgun metagenomic next-generation sequencing (smNGS) permits unbiased sequencing of DNA in a sample. Lately, there is excitement about the application of smNGS to infectious diseases as a diagnostics and research tool [16]. This partly stems from the decreasing cost of next-generation sequencing. smNGS has advantages over standard techniques, namely, bacterial cultures or quantitative PCR (qPCR), for the detection of microbes [17]. smNGS can sensitively detect and quantify the nucleic acid of any organism permitting the identification of multiple organisms in one sample. The sample-to-answer metagenomic box permits the disruption of the clinical infectious disease paradigm which relies on the physician formulating a differential diagnosis followed by sequential laboratory testing [18]. This traditional approach is challenged by overlapping clinical features of infectious causes and the non-existent tests for rare pathogens. Thus, a cause for IC/BPS cases was not been identified in patients for many years. IC/BPS is thought to be idiopathic and/or autoimmune in nature [19]. Ill- timed diagnosis in these patients lead to poor outcomes, increased anxiety, and high health care costs [20].
IC/BPS is a urologic chronic pelvic pain associated disorder with lower urinary tract symptoms with different potential etiologies. However, precise subtypes and phenotypes of IC/BPS remains unknown. In this study, we have employed smNGS to understand infection as a potential association and/or cause of IC/BPS due to its power of identifying a broad range of pathogens in a single test (Figure 1).
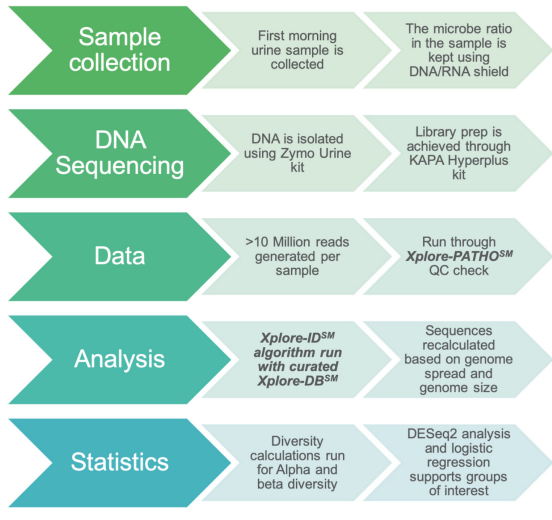
Figure 1: Shotgun metagenomics next-generation sequencing profiling workflow contains five major steps: sample collection, library preparation, sequencing, and data analysis along with statistics. The Xplore-Patho® analysis consists of six analysis modules. Xplore-Lib™ extracts custom genome reference libraries, Xplore-Map™ aligns the reads to the reference library and filters host reads, Xplore-ID™ identifies and estimates the proportions from each genome, and Xplore-Report™ provides detailed summary reports of the results. Xplore-DB™ provides additional annotation and Xplore-Filter™ is used to preprocess the reads prior to alignment.
Methods
Patients and Controls
All methods were carried out in accordance with relevant guidelines and regulation. The Aperiomics ethics committee provided an IRB exempt status to the study on the basis that the laboratory procedures were performed in a clinical laboratory to improve the utility of metagenomic sequencing in patients with urological symptoms and that all data was deidentified. All experiments were conducted in the Aperiomics Molecular Laboratory, a Clinical Laboratory Improvement Amendments (CLIA-49D2181329) and College of American Pathologists (CAP-866213701)-accredited clinical laboratory. All patients signed informed consent forms and patient protected information (PHI) was kept secure and anonymous. Clean-catch urine samples were obtained, from May 2017 to November 2018, from patients with a clinical diagnosis of IC/BPS given by their healthcare providers. 76 urine samples were screened, of which 57 samples were eligible for this study. Of these, 8 of IC/BPS samples were eliminated due to a number of reasons including subsequent longitudinal samples from the same patient, non-American samples, or samples with another definitive cause of bladder discomfort. Among what was considered the IC/ BPS group (n = 49 IC/BPS), the clinical diagnosis was distributed into various categories, namely, interstitial cystitis (n = 27 IC/ BPS), other cystitis (n=15 IC/BPS), cystitis unspecified (n = 6 IC/BPS), or acute cystitis (n = 1 IC/BPS). The gender distribution of the IC/BPS group was 8 males and 41 females. Thirteen out of the 15 controls were included in the study because they self-reported not having had a medical history of UTI, chronic diseases and no other genitourinary tract problems, with 9 female control samples and 4 male controls. The control patient urine sample cohort was selected based on self-reported absence of urinary tract infection health issues or recurrent/chronic pelvic pain. All samples were collected in sterile urine cups and Urine Conditioning Buffer (Zymo Research, Irvine, CA) was added immediately after collection to stabilize nucleic acids. Samples were shipped at ambient temperature and then stored at 4oC until processed for sequencing.
DNA Extraction, Library Preparation and Sequencing
Total DNA was isolated from each urine sample using the Quick-DNA Urine Kit (Zymo Research, Irvine, CA), per the manufacturer’s instructions. Libraries were prepared using the KAPA HyperPlus Prep Kit (KAPA Biosystems, Wilmington, MA). Quality control and quantification of the samples was assessed by fluorometry using the Qubit 2.0 Fluorometer (Thermo Fischer, Waltham, MA). DNA Libraries were sequenced on NextSeq500 (Illumina, San Diego, CA) using paired-end parameters (2x75 bp).
Quality Assessment
A minimum of 10 million reads was required for each sample. The quality of raw DNA sequencing reads was assessed using FastQC (v 0.11.8). An average per base sequence quality score of every sequence was determined and lower quality sequences and regions were trimmed when quality threshold scores fell below a Phred score of 25. The remainder of the sequences with average quality assessment scores below 25 or the trimmed read with length less than 35 base pair were excluded.
Analysis
All filtered sample data were run through Xplore-Patho® (Aperiomics, Inc., Sterling, VA), a proprietary metagenomic data analysis software based on a Bayesian sequencing read reassignment method that has been described before. Species were called when at least 30 reads mapped to a reference genome and were evenly distributed. Relative abundance of each species was accumulated as sequencing reads mapped to reference genomes, and species with less than 0.1% of the relative abundance were excluded from downstream analyses.
ANOVA and t-test statistical analyses were performed using Animalcules v0.99.71 in R (v3.6.0) shiny app for statistical microbiome analysis and plotted using Plotly and ggplot.
Age and gender were all evaluated for their predictive factors on the dataset to rule out covariates of low predictive value on the population. Gendered samples were evaluated separately as gender was determined to be a strong indicator of alpha-diversity and microbial composition as they align with literature and pilot studies.
Diversity Analysis
Alpha-diversity was calculated across patients and controls using Shannon and Gini-Simpson indices using Vegan v2.5-5. Differences in alpha-diversity between cases and controls were tested using the Welch t-test for samples with unequal variances. Bray-Curtis dissimilarity and Jaccard index to confirm Bray- Curtis results were observed looking for any differences between female IC/BPS and female controls. Bray-Curtis Beta-diversity was calculated using Bray-Curtis and evaluated with the Kruskal- Wallis ANOVA test.
Differential Analysis
Differential expression analysis of individual species level biosignatures was performed by DESeq2 looking at the Log2 fold changes and p values within the R package animalcules; adjusted p values were evaluated using the Independent Hypothesis Weighting method in DESeq2. DESeq2 is typically used for RNA seq data but has been shown to be useful for microbiome data concerning OTUs. The DNA data was normalized to the species genome size and converted to microbe counts per million for DESeq2 analysis [21,22].
Importance Plot
Phyla level importance plots were generated using logistic regression and random forests in R. For both tests the R package caret was used with the Generalized Linear “glm” and classic Random forest “ranger” models. Cross validation was done for both importance models using 10 folds and 1000 repeats.
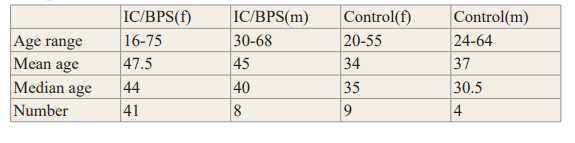
Table 1: Demographic information of our Patient and Control Samples, Including Gender and Age. The Largest Group Represented is Female. The IC Patients were on Average Older than the Controls. Various Analytic Tools Determined that Age was a Minor Factor in Microbiome Composition or Diversity.
Results
A total of 76 urine samples were screened, of which 57 samples were eligible and consented for this study. Of these, 8 of IC/BPS samples were eliminated due to a number of reasons including subsequent longitudinal samples from the same patient, non- American samples, or samples with another definitive cause of bladder discomfort. Among what was considered the IC/BPS group (n = 49 IC/BPS), the clinical diagnosis was distributed into various categories, namely, interstitial cystitis (n = 27 IC/BPS), other cystitis (n=15 IC/BPS), cystitis unspecified (n = 6 IC/BPS), or acute cystitis (n = 1 IC/BPS). The gender distribution of the IC/BPS group was 8 males and 41 females. Moreover, the female IC/BPS patient ages ranged from 16 to 75, with a median of 44, whereas, the male IC/BPS ages ranged from 30 to 68, with a median of 40 (Table 1). In contrast to the patient samples, 13 out of the 15 controls were included in the study because they self- reported not having had a medical history of UTI, chronic diseases and no other genitourinary tract problems, with 9 female control samples and 4 male controls. Female controls ages ranged from 20 to 55 with a median of 35 whereas male controls ages ranged from 24 to 64, with a median of 30.5 (Table 1).
Diversity
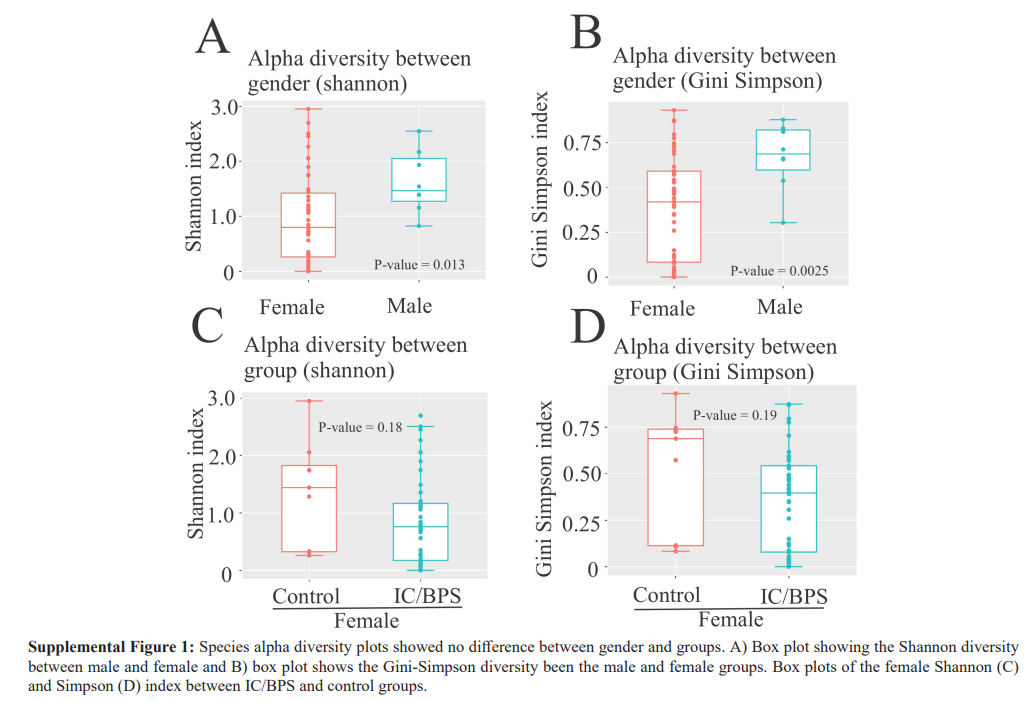
Generally, richness is an indicator of the overall number of different taxa regardless of distribution. Specifically, α-diversity refers to the diversity within a particular area or ecosystem, and is usually expressed by the number of species (i.e., species richness) in that ecosystem, while β-diversity refers to the ratio between local or alpha diversity and regional diversity. This is the diversity of species between two habitats or regions. We noted statistically significant differences between males and females through Shannon (p = 0.013) and Gini Simpson indices α-diversity (p = 0.0025, Supplemental Figure 1A, B). Due to α-diversity gender differences, this study focused on females since males had a distinct microbial composition and were fewer in number. In addition, α-diversity using the Shannon index was not significantly different between female control and female IC/BPS groups (p = 0.18; Supplemental Figure 1C, D).
Moreover, female control and female IC/BPS group comparisons found β-diversity differences (data not shown). Specifically, β-diversity was assessed by the Jaccard index and the Bray-Curtis dissimilarity. A significant difference in β-diversity between the female IC/BPS and control groups was found using the Kruskal- Wallis ANOVA (p = 2.33e-14) for nonparametric data.
Taxa Levels of Importance and Trends
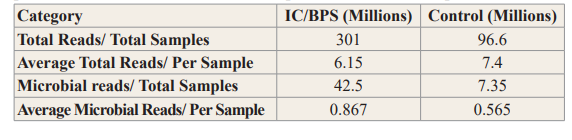
The IC/BPS and control sample sequencing statistics are shown in Table 2. Altogether, the total number of reads for each group was 301and 96.6 million reads, respectively. On average, each sample had 6.15 and 7.4 million reads for the IC/BPS and control groups, respectively. The total number of microbial reads for each cohort was 42.5 and 7.35 million reads for IC/BPS and control groups, respectively, and average microbial reads per sample were 867,000 (IC/BPS) and 565,000 (IC/BPS) reads.
Table 2: IC/BPS and Control Sample Sequencing Statistics. The Data are provided for the Total Batch of Sample and on a Per Sample Basis.
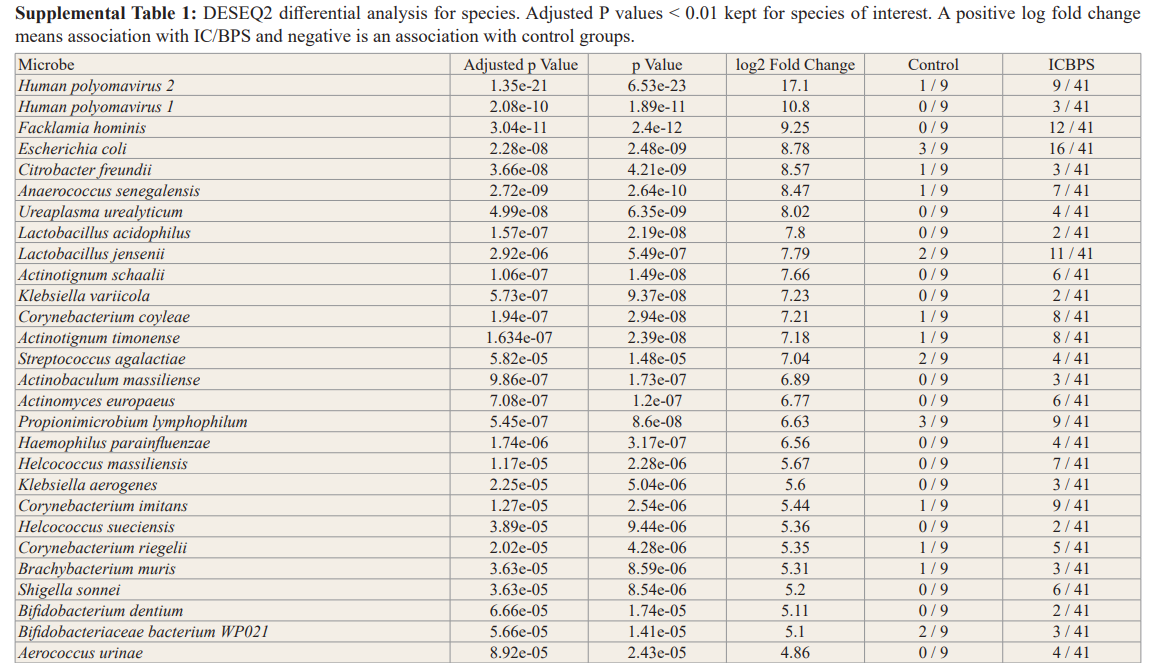
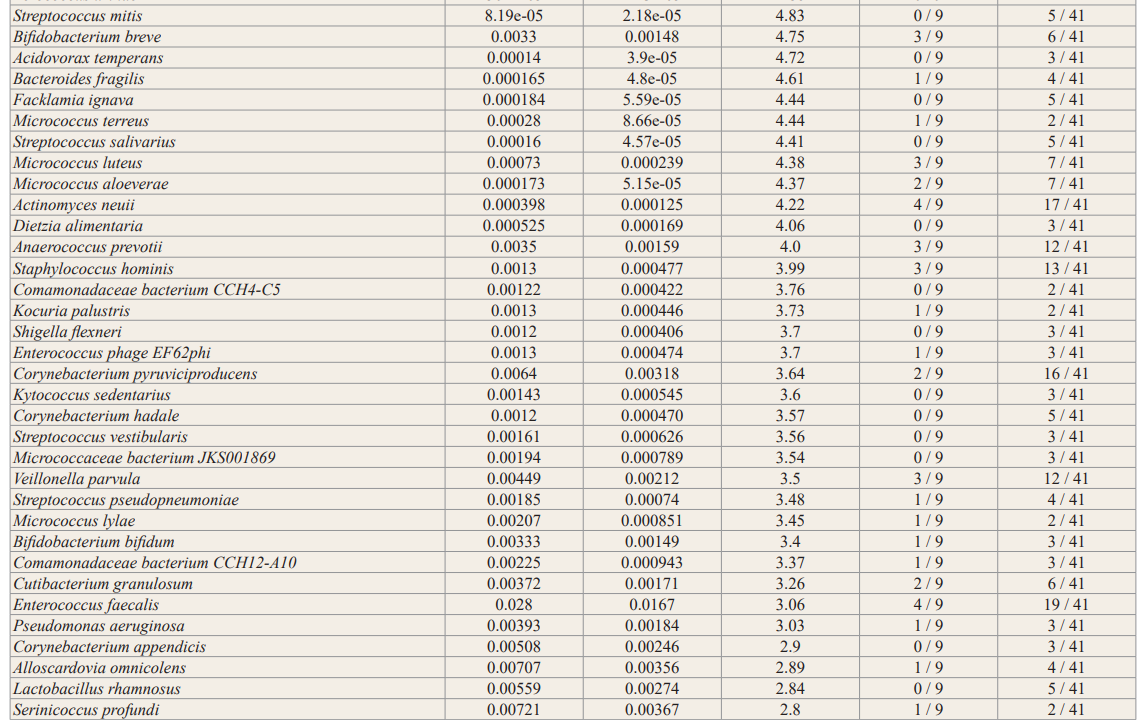
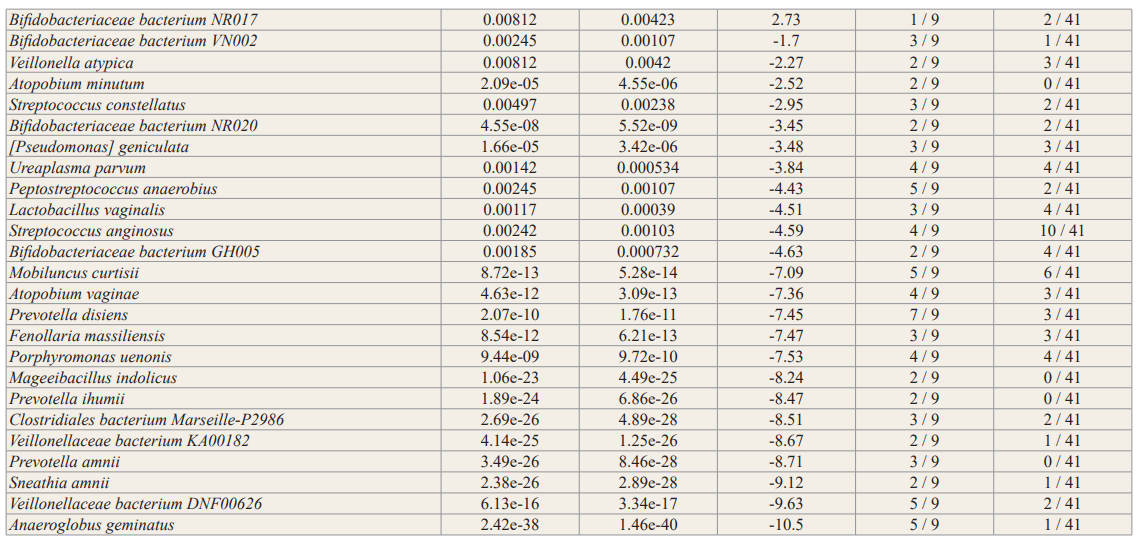
In general, no significance microbial load differences were observed between the female IC/BPS and control groups. Many of the female IC/BPS subjects had higher abundances of a number of specific groups of pathogens compared to control groups (Figure 2, Figure 3). Based on the female logistic regression analysis, the most significance phylum was the Proteobacteria (49% importance) (Figure 2, 3A), which contained the most common UTI bacteria E. coli. Random forest analysis also supported this finding (Figure not shown). The female AUC curve demonstrated that 71.7% of samples could be distinguished as IC/BPS or control based on Proteobacteria, Bacteroidetes, Firmicutes and Temecutes. Meanwhile, we noticed that BK and JC viruses were notably associated with the IC/BPS female group (Figure 3B). This may be partly explained by the small genome size of viruses because the relative abundances were calculated based on genome size to approximate the percentage of bacterial, fungal, and viral units present in the sample. In addition to bacteria and viruses, higher levels of fungi were identified in the IC/BPS group (Figure 3C). The yeast species that were found in the IC/ BPS group included Candida glabrata and Candida albicans. The filamentous fungi that were enriched in the female IC/BPS group included Apergillus spp., Penicillium rubens and Alternaria alternata. Aspergillus costaricaensis and Aspergillus fumigatus were only detected in male patients. The most abundant species in female IC/BPS patients were Lactobacillus iners and Lactobacillus crispatus. Similarly, the most abundant species in males was the JC virus (Figure 3D). In general, Lactobacillus, JC and BK viruses, and E. coli were observed to be more abundant in the female IC/ BPS group (Figure 3D). A detailed listing of the bacterial and viral species that are statistically significantly associated the IC/BPS are in Supplemental Table 1. A positive log-fold change signified an association with IC/BPS compared to control groups.
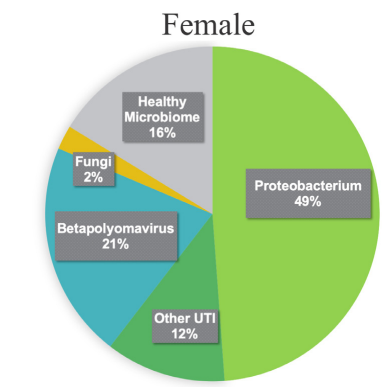
Figure 2: Most of the IC/BPS patients had virulence signatures and higher abundance levels of these microbes compared to control groups. The starkest of which was the Proteobacteria, which contains the most common UTI bacteria, E. coli. Other notable finds were the higher abundances of fungi and betapolyomavirus, of which standard urine testing cannot identify, whereas mNGS can.
Discussion
The etiology of IC/BPS remains unknown and the the microbial contribution unclear. Our results support the hypothesis that infection of either bacterial, viral, and/or fungal sources contributed to the IC/BPS symptoms in our study cohort (Figure 2 and 3). Based on traditional cultures, IC/BPS patients may remain undiagnosed and untreated for years. For example, E. coli, which was present in our IC/BPS group, can be easily cultured whereas other pathogens may not (Figure 2 and 3). A study evaluated
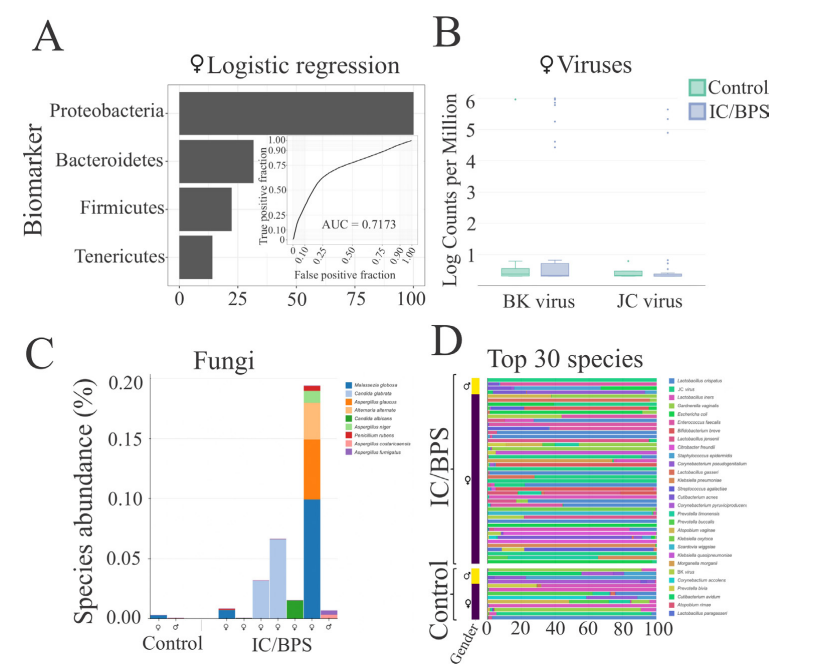
Figure 3: A) Logistic regression showed that phyla biomarkers are important, Proteobacteria was the strongest indicator. The AUC curve showed in the inner plot that 71.7% of the identifiers could be grouped as IC or control based on these phyla alone. B) Relative abundance was calculated based on aligned sequencing reads and genome size to approximate the percentage of bacterial, fungal, and viral units present in the sample. Because of this, the BK and JC polyomaviruses are notably visible. The vast majority of the remaining units on the chart are bacterial. C) Fungi are in all groups but are much more likely to be found in high abundance in the IC/BPS group. D and E) Bacteria from the phylum Proteobacteria and polyomaviruses are enriched in the IC/BPS group compared to the control group.
Urinary cultures in 428 IC/BPS patients and only 21.6% had colony forming units (CFU) from mostly Enterococcus, E. coli, Group B Strep and Klebsiella [23]. In contrast to the culturing approach, 33 IC patients were tested by 16S rRNA and detected bacterial DNA in 21/33 (64%) bladder samples [24]. Similarly, RFLP analysis identified E. coli DNA in 10/33 (30%) of the IC biopsy samples [24]. Moreover, 16S rRNA bacterial amplicons were found in 29% of patients (bladder samples) with IC [25]. Positive bacterial 16S rRNA amplification of bladder biopsy was seen in 16/30 (53%) IC patients [26]. Amplification of 16S rRNA from 6 IC patients and 6 controls from bladder biopsies was observed, which suggested the presence of E. coli; however, viable bacteria could not be cultured [27]. These results suggest the role of infection in IC/BPS, but other mechanisms cannot be ruled out, namely, inflammation and urothelial permeability associated with the urgency and frequency of pain.
To demonstrate the contribution of the microbiome to the disease, we employed standard analytical methods, namely, logistic regression for phyla microbial signatures, viral box-plots, fungal and bacterial stack bar graphs, alpha/beta diversity and DESeq2 differential relative abundance analysis to identify the similarities and differences between IC/BPS and control groups (Figures 2,3, Supplemental Figure 1, Supplemental Table 1). Outside of these most commonly studied species, the female IC/BPS group was associated with less commonly tested UTI species. The uropathogen Citrobacter freundii, a Proteobacteria, was highly associated with the IC/BPS group (Figure 3A, Figure 3D) [28]. UTIs caused by Citrobacter species were described in 5 to 12% of bacterial urine isolates in adults with UTI.
Standard UTI panels frequently miss some species implicated in infections. In our study, several samples had less common pathogens, for example, Ureaplasma urealyticum and Scardovia wiggsiae (Supplemental Table 1). We observed a female IC/BPS association with BK and JC viruses, which have been linked to rejection of kidney implants, prostatitis and progressive multifocal leukoencephalopathy (Figure 3B). Various Papovaviridae were associated with benign papilloma and carcinoma and through these growths, these viruses may cause hemorrhagic cystitis. Moreover, the BK polyomavirus family, was correlated with the clinical diagnosis or cytoscopic findings in IC/BPS patients; BK virus titers were found in 11 out of 15, with bladder ulcerations in 9 and the BK virus may be a urinary biomarker [29]. Although only one of the patients in this study’s IC/BPS group had been diagnosed with hemorrhagic cystitis, it is possible that other patients, that were BK or JC virus positive, had a low-level hemorrhage that was not tested for or undetected.
After a comprehensive characterization of all microbes within the urinary microbiome, fungi were identified at a lower level, but were highly associated with IC/BPS female and male groups (Figure 3C). A number of fungal taxa have been described in the urinary tract, by NGS sequencing of the internal transcribed spacer 1 (ITS1) region, in asymptomatic subjects [30]. However, like 16S sequencing for bacteria, this targeted amplicon sequencing technology does not have species level resolution. In contrast to healthy individuals, Candida spp. are commonly found as urinary pathogens, but other invasive fungal species, such as Cryptococcus, Aspergillus, Mucoraceae, Histoplasma, Blastomyces, and Coccidioides, can also infect the urinary tract [14]. In fact, fungi were found in urine from patients with urological chronic pelvic pain syndromes by the Ibis T-5000 universal biosensor system [31]. Some fungi may be underrepresented because they are difficult to disrupt by current extraction methods [32]. This may be one of the reasons why fungal levels were usually below 0.5% in IC/BPS; however, they were only seen in controls below 0.01%. In total, nine fungal species were uniquely identified in the IC/BPS group in this study (Figure 3C). Consistent with these findings, a study associated urinary fungi with urinary symptom severity among women with IC/BPS. Specifically, 13 fungal species (8 genera) were identified in 504 urine samples from 202 females. Longitudinal analyses demonstrated higher fungal diversity (Mean Ratio 3.8, 95% CI 1.3- 11.2, p= 0.02) and a higher likelihood of observing any fungal species (OR = 5.26, 95% CI 1.1-25.8, p= 0.04) in high versus low urinary severity individuals. Taxa analysis demonstrated increased presence and relative abundance of Candida (OR = 6.63, 95% CI 0.8-58.5, p= 0.088) and Malassezia (only identified in 'high' urinary severity phenotype) for high versus low urinary symptoms. This analysis suggested the possibility that greater urinary symptom severity is associated with the urinary mycobiome in females with IC/BPS.
Lactobacillus was unclear as a potential infectious agent. A decrease in Lactobacillus, which has antimicrobial properties, in both OAB and IC/PBS patients compared to controls was commonly observed [12,14,17]. Furthermore, the absence of Lactobacillus acidophilus has been shown to correlate with higher pain scores and higher scores on the interstitial cystitis symptom index [12]. Consistent with published literature at the genus level, jensenii were found to be associated with IC/BPS (adjusted p < 0.01); however, L. iners and L. crispatus were present in both groups. These data suggest a line of communication between the urinary microenvironment and underlying afferent nerves that is likely mediated by the urothelium.
The most common microbial exposure of the urethra is through nearby translocation of the microbiomes from the skin, gastrointestinal tract, and vagina, whose species and alpha diversity is determined by the gross anatomical and physiological differences. In general, the top skin and vaginal microbiome indicators at the genus level showed high microbial transfer but these species were not associated with disease (adjusted p >0.05; Figure 3A). Specifically, the top gastrointestinal (GI) species, such as Bacteroides spp. such as E. coli and E. faecalis, transferred from the GI tract to the lower urinary tract in the IC/BPS group. smNGS is cost-effective test, on a per species basis, less intrusive than other alternatives, such as cystoscopy or culture, and more comprehensive in identifying the species of the microbiome. For the IC/BPS group, a comprehensive species identification with best currently representation of the microbiome was key in detecting associations of less well-known pathogens. In contrast to the numerous advantages of smNGS, one limitation is the slower testing speed, which is comparable to culture-based methods. However, faster methods such as MALTI-TOF are not as comprehensive in the number of microbes that can be detected in one test. In IC/BPS cases where chronic suffering has been ongoing for years (sometimes decades), such comprehensive smNGS testing is invaluable.
In summary, by employing smNGS we demonstrated the potential association and/or cause of IC/BPS due to its power of identifying a broad range of pathogens in a single test.
The possible role of bacteria, fungi and viruses in the pathogenesis of IC/BPS has been investigated by smNGS in the present study. Hereby, of the 41 female patients tested who were diagnosed with IC/BPS, 35 had signs of infection with a microbial signature for IC/BPS, including the presence of Proteobacterium, JC and BK viruses, and higher fungal loads. Testing involving smNGS provided a more complete snapshot of the lower urinary tract microbes and was thus more suitable than culture, PCR, 16S, or 18S methods as indicators of a urinary infection. Implementation of smNGS to identify pathogens in IC/BPS patients, previously missed by other methods, will enable clinicians to make timelier and targeted therapeutic decisions.
Declarations
Ethics Approval and Consent to Participate
Informed written consent was obtained for all subjects to participate in this study and data was deidentified. All experiments were conducted in the Aperiomics Molecular Laboratory (AML), a Clinical Laboratory Improvement Amendments (CLIA- 49D2181329) and College of American Pathologists (CAP- 866213701)-accredited clinical laboratory. This study was exempt from requiring ethics approval on the basis of deidentified data and data was obtained within a clinical laboratory environment to improve the utility of metagenomic sequencing in patients with urological symptoms.
Availability of Data And Materials
The datasets generated and/or analysed during the current study are available in the BioProject repository (Accession: PRJNA679884 ID: 679884) at https://www.ncbi.nlm.nih.gov/bioproject/679884.
Authors' Contributions
AWL: Data analysis, Manuscript Writing, Read and approved final manuscript. CAV: Study Design, Data analysis, Manuscript Writing, Read and approved final manuscript. YC: Analysis pipeline design and execution, Data analysis, Manuscript Editing, Read and approved final manuscript. KAC Analysis pipeline design and execution, Manuscript Writing, Read and approved final manuscript. CRI: Data collection or management, Manuscript Writing, Read and approved final manuscript.
References
- Van de Merwe JP, Nordling J, Bouchelouche P, et al. Diagnostic criteria, classification, and nomenclature for painful bladder syndrome/interstitial cystitis: an ESSIC proposal. Eur Urol. 2008; 53: 60-67.
- Wennevik GE, Meijlink JM, Hanno P, et al. The Role of Glomerulations in Bladder Pain Syndrome: A Review. J Urol. 2016; 195: 19-25.
- Hughes PA, Zola H, Penttila IA, et al. Immune activation in irritable bowel syndrome: can neuroimmune interactions explain symptoms?. Am J Gastroenterol. 2013; 108: 1066-1074.
- Contreras-Sanz A, Krska L, Balachandran AA, et al. Altered urothelial ATP signaling in a major subset of human overactive bladder patients with pyuria. Am J Physiol Renal Physiol. 2016; 311: F805-F816.
- Balachandran AA, Wildman SS, Strutt M, et al. Is chronic urinary infection a cause of overactive bladder? Eur J Obstet Gynecol Reprod Biol. 2016; 201: 108-112.
- Khasriya R, Sathiananthamoorthy S, Ismail S, et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol. 2013; 51: 2054-2062.
- Walsh CA, Siddins A, Parkin K, et al. Prevalence of “low- count” bacteriuria in female urinary incontinence versus continent female controls: a cross-sectional study. Int Urogynecol J. 2011; 22: 1267-1272.
- Gill K, Kang R, Sathiananthamoorthy S, et al. A blinded observational cohort study of the microbiological ecology associated with pyuria and overactive bladder symptoms. Int Urogynecol J. 2018; 29: 1493-1500.
- Wood MW, Breitschwerdt EB, Nordone SK, et al. Uropathogenic E. coli promote a paracellular urothelial barrier defect characterized by altered tight junction integrity, epithelial cell sloughing and cytokine release. J Comp Pathol. 2012; 147: 11-19.
- Abraham SN, Miao Y. The nature of immune responses to urinary tract infections. Nat Rev Immunol. 2015; 15: 655-663.
- Vijaya G, Cartwright R, Derpapas A, et al. Changes in nerve growth factor level and symptom severity following antibiotic treatment for refractory overactive bladder. Int Urogynecol J. 2013; 24: 1523-1528.
- Curtiss N, Balachandran A, Krska L, et al. A case controlled study examining the bladder microbiome in women with Overactive Bladder (OAB) and healthy controls. Eur J Obstet Gynecol Reprod Biol. 2017; 214: 31-35.
- Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014; 52: 871-876.
- Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio. 2014; 5: e01283-e01314.
- Abernethy MG, Rosenfeld A, White JR, et al. Urinary Microbiome and Cytokine Levels in Women with Interstitial Cystitis. Obstet Gynecol. 2017; 129: 500-506.
- Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019; 20: 341-355.
- Hilton SK, Castro-Nallar E, Pérez-Losada M, et al. Metataxonomic and Metagenomic Approaches vs. Culture- Based Techniques for Clinical Pathology. Front Microbiol. 2016; 7: 484.
- Greninger AL, Naccache SN, Federman S, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015; 7: 99.
- Van de Merwe JP. Interstitial cystitis and systemic autoimmune diseases. Nat Clin Pract Urol. 2007; 4: 484-491.
- Khetsuriani N, Holman RC, Anderson LJ. Burden of encephalitis-associated hospitalizations in the United States, 1988-1997. Clin Infect Dis. 2002; 35: 175-182.
- Sophie Weiss, Zhenjiang Zech Xu, Shyamal Peddada, et al. Normalization and Microbial Differential Abundance Strategies Depend Upon Data Characteristics. Microbiome. 2017; 5: 27.
- McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014; 10: e1003531.
- https://www.ics.org/2018/abstract/57?gclid=Cj0KCQjw6sHzBRCbARIsAF8FMpU-Dr7zlLUVJicEsLnRKuhdqtzH5MlFq dqRhxJZBbttQX-FuvPowQkaAoefEALw_wcB.
- http://ispub.com/IJU/7/1/5137.
- Domingue GJ, Ghoniem GM, Bost KL, et al. Dormant microbes in interstitial cystitis. J Urol. 1995; 153: 1321-1326.
- Heritz DM, Lacroix JM, Batra SD, et al. Detection of eubacteria in interstitial cystitis by 16S rDNA amplification. J Urol. 1997; 158: 2291-2295.
- Keay S, Zhang CO, Baldwin BR, et al. Polymerase chain reaction amplification of bacterial 16S rRNA genes in interstitial cystitis and control patient bladder biopsies. J Urol. 1998; 159: 280-283.
- Ranjan KP, Ranjan N. Citrobacter: An emerging health care associated urinary pathogen. Urol Ann. 2013; 5: 313-314.
- Grigorescu B, Powers K, Lazarou G. Update on Urinary Tract Markers in Interstitial Cystitis/Bladder Pain Syndrome. Female Pelvic Med Reconstr Surg. 2016; 22: 16-23.
- Ackerman AL, Underhill DM. The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann Transl Med. 2017; 5: 31.
- Nickel JC, Stephens A, Landis JR, et al. Assessment of the Lower Urinary Tract Microbiota during Symptom Flare in Women with Urologic Chronic Pelvic Pain Syndrome: A MAPP Network Study. J Urol. 2016; 195: 356-362.
- Tang J, Iliev ID, Brown J, et al. Mycobiome: Approaches to analysis of intestinal fungi. J Immunol Methods. 2015; 421: 112-121.