Expression of NF-kB Regulators (BCL-3, NFKB1, NFKB2) in Metastatic Breast Cancer Patients
Author'(s): Arta Fejzullahu1*, Tugba Akin Telli2, Irem Peker Eyuboglu1, Perran Fulden Yumuk2,3 and Ahmet Ilter Guney4
1Department of Medical Biology and Genetics, Institute of Health Sciences, Marmara University, Istanbul, Turkey.
2Division of Medical Oncology, School of Medicine, Marmara University, Istanbul, Turkey
3Department of Medical Oncology, School of Medicine, Koc University, Istanbul, Turkey.
4Department of Medical Genetics, School of Medicine, Marmara University, Istanbul, Turkey.
*Correspondence:
Arta Fejzullahu, Department of Medical Biology and Genetics,Institute of Health Sciences, Marmara University, 34854,Istanbul, Turkey, Tel: +90 505 144 96 17.
Note: Arta Fejzullahu is still a faculty member of Istanbul Aydin University.
Received: 14 May 2021; Accepted: 24 June 2021
Citation: Fejzullahu A, Telli TA, Eyuboglu IP, et al. Expression of NF-KB Regulators (BCL-3, NFKB1, NFKB2) in Metastatic Breast Cancer Patients. Cancer Sci Res. 2021; 4(2): 1-9.
Abstract
Background/aim: BCL-3, upregulated in several cancers, functions as a crucial player not only in cell cycle and apoptosis, but also in metastasis. The study aimed to analyse the expression profile of BCL-3 and its interacting partners in metastatic breast cancer.
Materials and methods: mRNA expression levels were evaluated in blood samples of metastatic breast cancer patients (n=55) and in healthy control donors (n=50) by RT qPCR. SPSS 26 program was used for statistical analysis.
Results: Expression levels of BCL-3 mRNA was downregulated in metastatic breast cancers patients compared to healthy control group (p= 0.0004). Subsequently, expression levels of the NFKB1 (p= 0.0005), NFKB2 (p=0.0001), CYLD (p= 0.00042) and TP53 (p= 0.0287) were significantly lower in metastatic breast patients compared to healthy controls. CCND1 and CDH2 gene expressions were significantly upregulated in metastatic group (p=0.0009; and p= 0.0030, respectively). GSK3B, SMAD3 and TGFB1 expressions were not significantly different between groups.
Conclusion: This study indicates significant expression difference between patients and controls. The NF-κB regulators might induce the metastatic potential through interaction with the other molecules in the microenvironment, nevertheless, it is needed further research whether the importance of BCL-3 as a biomarker for metastatic breast cancer.
Keywords
Introduction
Breast cancer is the leading cause of cancer-related deaths worldwide in women with more than 630 thousand deaths per year [1]. Failure to fully treat advanced breast cancer can be attributed to its being systemic disease after metastasis, which shortens patient’s survival rate and unfortunately results in undesirable health conditions [2].
Recent advances in anti-hormonal therapy, targeted drugs and immunotherapeutic interventions have improved treatment response rates in specific subtypes (hormone receptor positive,
HER-2-enriched), but even more effective therapies are urgently needed to achieve a significant improvement in the survival outcome of patients especially for more aggressive triple negative breast cancer (TNBC) subtype [2-3]. Better understanding of tumour’s molecular biological features and faster translation of current knowledge into clinically useful tools wait to be met for metastatic patients and the scientific community alike [4].
In this respect, many signalling pathways have been studied in order to offer insight into pathogenesis and progression of metastatic breast cancer searching for prognostic and predictive markers, which can be used in diagnosis and treatment for many years. Recently, a promising mediator of NF-κB signalling pathway B-cell lymphoma 3 (BCL-3), a proto-oncogene candidate, has been shown as the therapeutic target due to its significant role in cell death and proliferation [5]. BCL-3, a nuclear atypical member of IkB (inhibitor of NF-κB) family, by interacting with NF-κB p50 (transcribed by NFKB1) and p52 (transcribed by NFKB2) homodimers, could act as an activator or repressor of NF-κB transcription factors, which exert their function through classical and alternative pathways (Figure 1) [4,6,7].
Beside BCL-3’s proliferative and apoptotic role, recent studies have shown that BCL-3 gene might also be associated with development of metastasis, and inhibition of BCL-3 may reduce metastasis. Puvvada et al. [8] demonstrated that nuclear expression of BCL-3 is strongly associated with survival in metastatic colorectal cancer, making it an interesting prognostic marker [8].
Association between BCL-3 and metastatic breast cancer has been previously highlighted in a HER2-positive mouse tumor model, and this phenomenon was suggested to be involved in promoting metastasis of BCL-3 but did not affect primary tumor growth [9]. The role of BCL-3 in promoting metastasis has been independently associated with an increase in cell motility and has recently been linked to regulation of TGF-signaling through stabilization of SMAD3 [10]. Additionally, several studies showed that elevated BCL-3 expression may activate invasion and metastasis in different types of cancer [breast, colorectal, gastric, non-small-cell lung cancer] [7,9,11]. All these data suggest that BCL-3 may also promote cell migration and metastasis. Another important study has also shown that excessive BCL-3 gene expression increases resistance to cytotoxic effect of drugs in people undergoing chemotherapy [12]. Accordingly, instead of global NF-κB signal inhibition, which may lead to non-target toxicity, targeting BCL-3 gene with novel therapeutic drugs that specifically serve on the NF-κB pathway, may be a promising route, which deserves to be studied further in metastatic breast cancer.
In this line, we will evaluate the expression of major intracellular regulators of NF-κB pathway at mRNA level in fresh blood samples of metastatic patients. Purpose of this study was to compare the gene expression of NF-κB regulators (BCL-3, NFKB1, NFKB2) and other BCL-3 interacting genes (CYLD, GSK3B, CCND1, TP53, CDh3, SMAD3, TGFB1) between metastatic breast cancer patients and healthy controls by Reverse Transcriptase- Quantitative Real-Time PCR (RT-qPCR).
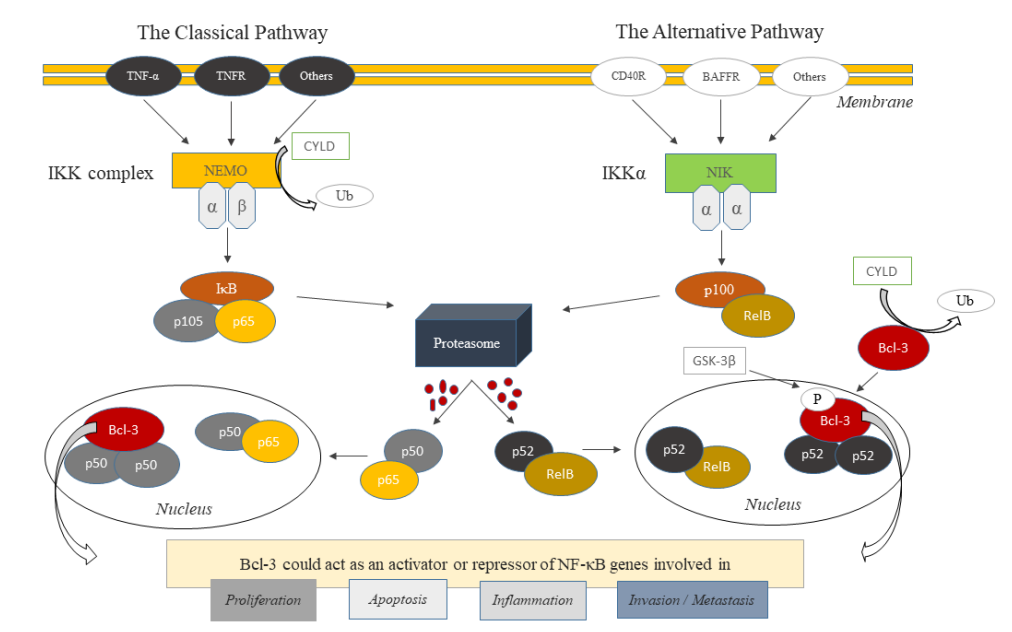
Figure 1: Classical and Alternative NF-κB Signaling Pathways (modified from reference 4).
Material and Methods
Study design, population, and blood specimens and data collection. In this study, ethical guidelines of the Helsinki Declaration were followed [13]. Prior to study initiation, approval was granted by the Scientific Committee and the Committee on Research and Ethics of the Marmara University [Turkey]. Informed consent was obtained from all the participants.
mRNA expression of three members of the alternative pathway (BCL-3, NFKB1 and NFKB2) and other genes interacting with were studied in metastatic breast cancer patients (n=55, unrelated Turkish women) who were followed up at Medical Oncology Division of Marmara University since 2015. Patients were actively treated and demographic data, tumor characteristics, and treatment details and were retrospectively reviewed from their files.
Data obtained from patients was compared with healthy control donors. Control group (n=50) was selected from healthy donors of Medical Genetics Department of Marmara University and Istanbul Aydin University academic staff. Members of the control group did not have any history of chronic inflammatory diseases, were free from metastatic breast cancer and/or any other cancer type at the time of the study and their functional capacity was sufficient. Workflow of the study is described in Figure 2.
Gene Expression Analysis by Reverse Transcriptase- Quantitative Real-Time PCR (RT-qPCR)
RNA preparation
Fresh blood specimens from 55 metastatic breast cancer patients and 50 controls were used to extract RNA samples. Roche RNA isolation kit for mammalian blood [Roche, Munich, Germany], which is commercially available, was used according to the manufacturer’s instructions. Total RNA was quantified using a NanoDrop 1000 Spectrophotometer (Fisher Thermo, Wilmington, DE, USA), and was stored at −80°C. All sample processing was performed at Medical Genetics Department of Marmara University Hospital.
cDNA synthesis
Total of 1 μg of RNA was reverse transcribed into cDNA using 100U of iScriptTM cDNA Synthesis Kit [Bio-Rad] following the manufacturer’s instructions in a total volume of 20 μl. Kit includes all reagents (5x iScript Reaction mix, iScript Reverse Transcriptase and Nuclease-free water) required for reverse transcriptase. As a control of nucleic acid contamination, a sample (RNA) free negative control was used in each run. The mixture was incubated in a C1000 Touch thermal cycler (Bio-Rad) at 25°C for 5 minutes (priming), 40°C for 20 minutes [reverse transcription], and 95°C for 1 minutes (RT inactivation). Accordingly, reverse transcription products (cDNA products) were diluted to a concentration such that the Ct (cycle threshold) value of the diluted solution was well within the range of detection capacity of the qPCR apparatus that was determined by a standard curve and stored at −20°C.
Primer Design
Specific primer sequences for amplifying coding regions for the functional domain of target genes and housekeeping control gene were designed using the primer designing tool Primer3 software and BLAST, according to the sequences provided in NCBI database. Primers were synthesized by OLIGOMER biotechnology. Primer sequences used in this study are listed in Table 1.
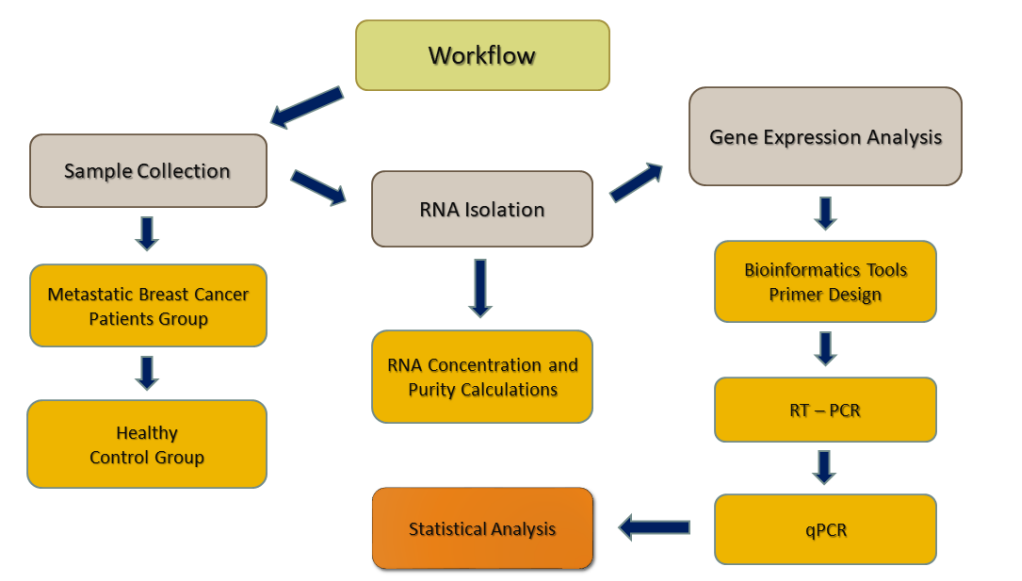
Figure 2. Overview of the workflow of the current study.

Quantitative Real Time-PCR (qPCR)
Expression levels of the BCL-3, NFKB1 and NFKB2 and other genes were quantified by qPCR assays. qPCR was carried out in CFX Connect Real-Time PCR Detection System. SsoAdvanced Universal SYBR Green Super mix (both from Bio-Rad) kit was used for performing qPCR reaction. Total volume of reaction mix (20 μl) includes SsoAdvanced Universal SYBR Green Super mix (2x), forward and reverse primers (250nM), Nuclease-free water and cDNA template (5μl). Cycling conditions were as follows: 30 seconds at 95oC [Polymerase Activation and DNA denaturation], 35 cycles of 95oC for 5 seconds (denaturation), 60oC for 15 seconds for annealing/extension (for all target genes).
Relative Quantification
Cycle threshold (Ct) is defined as PCR cycle number at which fluorescence generated from amplification of the target gene within a sample increases to a threshold value of 10 times the standard deviation of the base line emission and is inversely proportionate to the starting amount of the target cDNA. Relative quantities were corrected for efficiency of amplification and fold change in gene expression between groups was calculated. Relative expression was calculated via the comparative Ct method, 2−â??â??Ct for each sample, where â??Ct=Cttarget−Ctreference and ΔΔCt=â?³Ctpatient−â?³Cthealthy. Conditions for PCR and melting curves performed after amplification were tested and optimized, and thus the product specificity was checked by melting curve analysis and gene expression results were normalized to housekeeping control (reference) gene ACTB (beta actin) levels.
Statistical analysis
Statistical Package for Social Sciences Version 26 (SPSS, Chicago, IL, USA) and Graph Prism programs were used for statistical analysis. To test the distribution of data, Kolmogorov-Smirnov and Shapiro-Wilk analyses were performed and histograms were plotted. Metastatic breast cancer patients’ expression results, versus control group were compared. Student’s two-sided t-test was used to compare data between two groups. Mean â??Ct values and relative expression were compared with independent sample t-test. Difference was considered as statistically significant at p<0.05. While performing qPCR, statistical evaluations were made with ΔCt values and â??â??Ct method was used in order to calculate the relative fold change gene expression of samples.
Results
Clinical parameters and molecular characteristics of case-control group. This study included 55 metastatic breast cancer patients and 50 healthy control donors. Median age of the patients and healthy controls were 52 (range 31-81) and 27 (range 19-52) years, respectively. Study population (n=105) was Turkish in ethnicity and female in gender. De novo metastatic disease was seen in 45.5% of patients and all patients had metastasis at different locations (brain, bone, lungs, liver, lymph nodes, pleura) and were already receiving chemotherapy. Histological subtypes of patients were invasive ductal carcinoma (76.7%), invasive lobular carcinoma (1.8%) and other subtypes (21.8%). Study included Luminal A (30.9%), Luminal B (45.5%), HER2+ (14.5%) and TNBC(9%) molecular subtypes classified according to expression levels of ER, PR and HER2 protein status. Among total study population 15.3% of patients were smokers and 84.7% non- smokers. Forty percent of patients had family history of cancer and control group had no known cancer in their families. Clinical and molecular characteristics of the study population are summarized in Table 2.
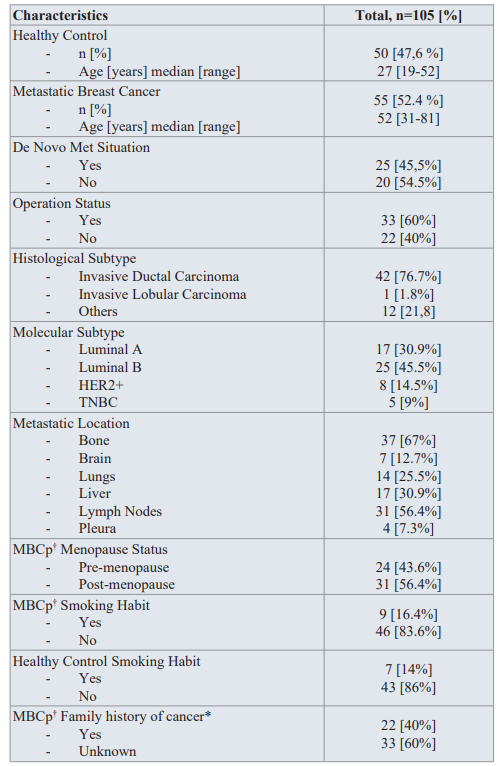
Table 2: Demographic data of case-control group.
† MBCp: Metastatic breast cancer patients
Elevated Expression of NF-κB Regulators in Metastatic Breast
Cancer Patients
Expression profile of NF-κB Regulators (BCL-3, NFKB1, NFKB2) and considered interacting partners of BCL-3 gene (CYLD, GSK3B , CCND1, TP53, CDh3, SMAD3, TGFB1) in patients (n=55) and healthy control donors (n=50) were examined to evaluate the role of BCL-3 gene in prognosis and diagnosis of metastatic breast cancer patients.
Respectively, BCL-3 expression was downregulated in metastatic breast cancer patients compared to healthy controls (fold change: 0.78; p <0.0004). Subsequently, expression levels of other two key regulators of NF-κB signaling pathway were significantly lower in metastatic patients compared to healthy controls (NFKB1: fold change 0.73; p = 0.0005 and NFKB2: fold change 0.68; p < 0.0001). Besides these regulators, we also determined the expression levels of BCL-3 considered interacting partners. Expression levels of CYLD, which was demonstrated to be involved in BCL-3 deubiquitylation was also found to be downregulated in metastatic breast cancer patients compared to healthy controls (fold change: 0.71; p = 0.00042). On the contrary, expression of GSK3B, considered to regulate BCL-3 phosphorylation, was not observed to be significantly different even it seemed to be downregulated in case group (fold change: 0.79; p = 0.0581). Expression of CCND1 gene, functioning in cell cycle progression, was upregulated in metastatic group and shows a significant p value (fold change: 1.61; p = 0.0009). Another important tumor suppressor gene, TP53, was observed to be downregulated (fold change: 0.85; p = 0.0287) in case group compared to control group. CDh3, an important gene with its role in cancer metastasis, was upregulated in metastatic breast cancer patients compared to healthy control group (fold change: 1.59; p = 0.003). Another two genes (TGFB1 and SMAD3) which were previously demonstrated to be associated with bone metastasis of breast cancer were also studied. However, neither SMAD3 nor TGFB1 expression was significantly different between case and control group (fold change; 0.8; p = 0.1770 and fold change: 0.92; p = 0.1324, respectively). Relative expression and p values of all studied genes in metastatic breast cancer patients and healthy control donors are shown in Figure 3.
Expression Levels in Molecular Subtypes and Clinical Parameters
Expression levels of all studied molecules were also analysed among molecular subtypes of metastatic breast cancer. Four molecular subtypes were Luminal A, Luminal B, HER2+ and TNBC. Firstly, we compared expression profile between four subtypes, then between each other. NFKB1 (p= 0.043) and NFKB2 (p= 0.042) expression profiles were observed to be significantly different among molecular subtypes. BCL-3 (p= 0.01), NFKB1 (p= 0.047) and NFKB2 (p= 0.007) gene expressions showed significant difference between Luminal A and B groups. Expressions for NFKB1 (p=0.027), CDh3 (p= 0.014) and CCND1 (p=0.021) were found to be significantly different between Luminal A and HER2+ groups. Expression levels of NFKB1 (p= 0.013) differed between Luminal A and TNBC molecular subtypes. Expression level of CYLD, GSK3B, TP53, SMAD3 and TGFB1 did not show any significant difference between molecular subtypes.
Then, we analysed expression significance in terms of clinical parameters (smoking habit, menopause status, operation status) among study population and also between case and control group, but no statistical difference (p>0.05) was detected in the expression profile of all genes studied.
Discussion
Association of NF-κB regulators and cancer was reported in several studies. NF-κB regulators exert their function through the activation of classical and alternative NF-κB signaling pathway. Two pathways are involved in several cellular activities such as cell cycle control, apoptosis and other processes. Among several intracellular components of NF-κB regulators [p105/p50 (NFKB1), p100/p52 (NFKB2), p65 (RelA), RelB, c-Rel, BCL-3] key players of pathways are NFKB1, NFKB2 and BCL-3 [4,6,14].
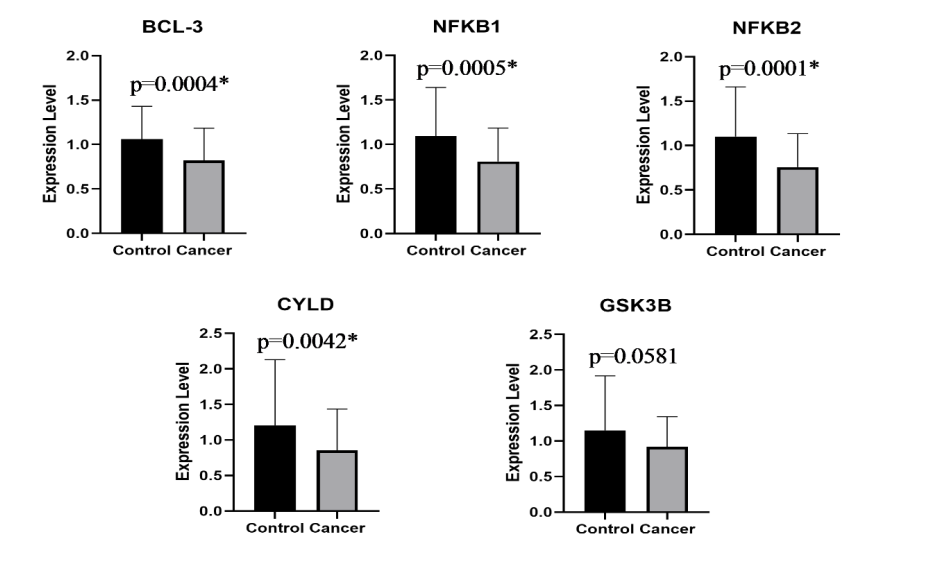
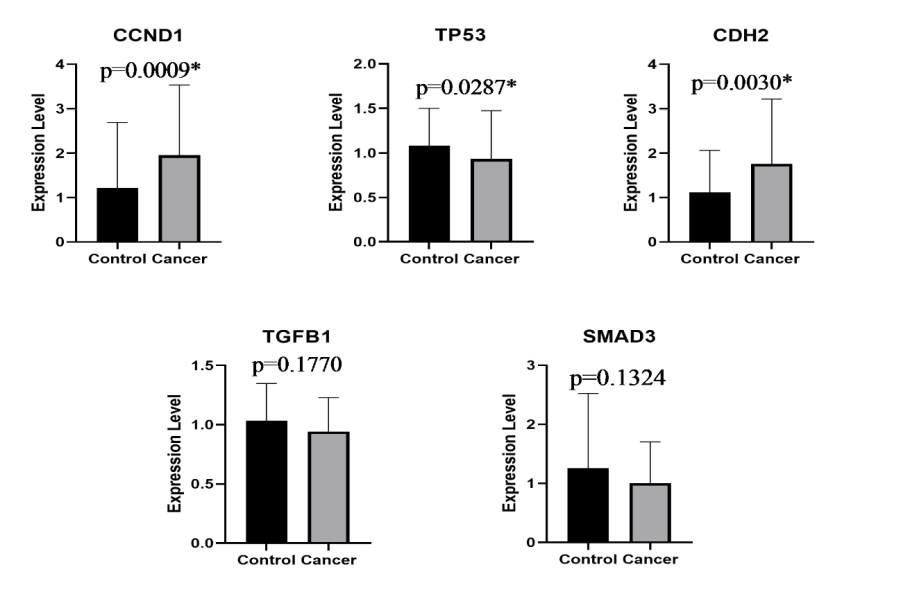
Figure 3: Relative expression in mRNA levels of all studied genes in metastatic breast cancer patients and healthy control donors.
*Indicates significant values, p<0.05.
In this study we especially focused on NF-κB signaling mediator BCL-3 gene expression and its considered interactive partners that were demonstrated to be involved in cancer progression [5,7,9- 11,14-24]. Studies of BCL-3 showed that alternated expression profile was involved not only in cell proliferation and in apoptosis, but also in metastasis in several cancer types and might have significant role as a key regulator in metastasis formation [8-11]. Since 20-30% of breast cancer patients may develop metastasis after diagnosis and primary tumor treatment, and approximately 90% of cancer-related deaths are attributed to metastasis, studies on BCL-3 gene have targeted on metastasis as a diagnostic or prognostic marker. Therefore, because there is a need to find and advance highly sensitive biomarkers that can be obtained from patients with non-invasive or minimally invasive methods BCL-3 may be used also as an non-invasive biomarker (due to its gene elevated expression in several cancers) in the future. Moreover, since NF-κB transcription pathway has a pivotal role in the regulation of immune system against cancer as well as in promoting inflammation and tumor development, the global inhibition of NF-κB signaling pathway could result in toxic effects. Therefore, BCL-3 targeted drugs might be as a promising route to stop cancer progression without toxic effects.
Human breast cancers display nuclear accumulation of BCL-3, as well as NFKB1 and NFKB2 regulators [22]. These activated molecules enhance cellular proliferation, cause decreased apoptosis and even might lead to cancer progression, but what triggers the activation of NF-κB regulators in breast cancer is still unknown [22]. Therefore, as this is our pilot study we started with non-invasive detection of the expression profile of BCL-3 and its’ considered partners in fresh blood samples of metastatic breast cancer patients. Analysis of blood samples may provide information if BCL-3 could be used as a non-invasive diagnostic biomarker. Moreover, to our knowledge, this is the first study that demonstrates the expression of BCL-3 and its interactive partners in Turkish population with multiple metastatic breast cancer patients.
In this study, mRNA expression profile of NF-κB regulators [BCL-3, NFKB1, NFKB2] have shown decreased gene expression in metastatic breast cancer patients when compared with healthy controls. In fact, we expected to observe increased expression due to BCL-3’s oncogenic feature previously demonstrated in nasopharyngeal, endometrial and prostate cancer studies [7- 10,17,21,24-25] as elevated expression of oncogenes generally are believed to promote cell proliferation and enhance chemo- resistance [7,24]. However, in accordance with our results there is another study that has shown repressed expression of BCL-3 in inflammatory bowel disease. The study determined commonly methylation in CpG sites of BCL-3, which explains the repression status [29]. However, because we have not analyzed the methylation status of BCL-3 in our patients we have to verify this downregulation in our further studies.
Moreover, when we compare our results with the literature, it is important to highlight that our patients were already receiving chemotherapy. In this respect, chemotherapy might affect or suppress expression level of our NF-κB regulators so we need to further analyses the expression profile of samples taken before and after chemotherapy since BCL-3 might act as an oncogene before chemotherapy. This reminds us of the study, which has shown that excessive BCL-3 gene expression increased resistance to cytotoxic effect of drugs in patients undergoing chemotherapy [12]. So decreased expression of BCL-3 might result the opposite and hence have a dual prognostic and predictive role in metastatic breast cancer. A previous study in TNBC cell lines (BL1 Subtype) have demonstrated that knockdown of BCL-3 gene could inhibit cell proliferation and elevates the sensitivity of patients and cell to chemotherapeutic agents [24]. Furthermore, NF- κB1 activation might be anti-apoptotic or pro-apoptotic depending on cell type or chemotherapy treatment stimulus. Beside breast cancer studies there is another study that have shown downregulated BCL- 3 expression in colon cancer due to drug treatment [30]. In this line as our case-control group size is limited further analysis is needed to determine chemotherapy effect on gene expression or drug resistance. To verify chemotherapy effect on expression levels we have to obtain larger number of each subgroup (Luminal A, Luminal B, HER2, TNBC) as breast cancer is not considered a single cancer type and the chemotherapy approach to each subtype is different. Since the treatments applied can change the expression of the genes investigated, it is crucial which treatments are given. Therefore, to overcome this problem our first goal as a continuation of this work will be to enlarge the patient group for each subtype and evaluate it according to the therapy they receive. Additionally, we have to verify our expression results in blood and tumor samples of our metastatic patients not only in mRNA level but also in protein level to obtain valuable results. Moreover, in subsequent work it should be better to compare our expression results with loco regional breast cancer disease not only with healthy controls to make a better assumption of role in metastatic disease since expression levels could also be increased in localized diseases.
Beside NF-κB regulators, we also analyzed gene expression of several genes that could interact with BCL-3. These genes were selected based on literature research on BCL-3 studies. In this line, one of the selected gene was CYLD. Downregulated CYLD that is required to execute BCL-3 deubiquitylation for preventing the nuclear accumulation of BCL-3, might cause an interaction with BCL-3 expression, and thereby promote cancer progression [5,14,26]. GSK3B gene, involved in BCL-3 phosphorylation was also downregulated, however did not show any statistical significance. We propose that GSK3B needs to be further studied since it is having been suggested that only phosphorylated BCL-
3 could exert a full effect in the DNA binding properties of NFKB1 homodimer [5-7,21]. Similarly, to previous data reported, overexpression of CCND1 was observed in metastatic breast cancer patients compared to healthy controls in our study demonstrated a strong correlation to metastatic situation [9,11,16]. Expression of TP53 gene was detected to be downregulated in metastatic breast cancer patients. Although 20-30% of mutations in TP53 occur in breast cancer, we did not analyze the whole gene for mutations [18,27]. In accordance to literature, overexpression of CDh3 was observed in metastatic breast cancer patients compared to healthy control group [28]. Another two genes (TGFB1 and SMAD3) that were demonstrated to be associated with bone metastasis of breast cancer by affecting tumor angiogenesis and epithelial- mesenchymal transition (EMT) were detected [10]. However, neither the expression of SMAD3 nor the expression of with TGFB1 was significantly different between study groups.
In conclusion, in this study we analyzed the expression levels of NF-κB regulators (BCL-3, NFKB1, NFKB2), and presented significant expression difference of studied genes in metastatic breast cancer patients with multiple metastatic locations. In this regard, the NF-κB regulators might induce the metastatic potential through interaction with the other molecules in the microenvironment, nevertheless, it is needed further research whether the importance of BCL-3 on breast cancer oncogenesis and as a biomarker of diagnosis and prognosis of metastasis in breast cancer patients. However, neither the molecular details of the interactions between BCL-3 and partner molecules are known, nor the BCL-3 mediated network of interactions is understood. It is obvious that this pilot study is only hypothesis generating. Hence, larger genomic, functional and pharmacogenomics studies before and after chemotherapy in tumor samples is needed to further assess diagnostic or prognostic role of BCL-3, identify its interactive partners, and the effect of chemotherapy on the expression levels of NF-κB regulators in each subtype of metastatic breast cancer patients in a larger group.
Acknowledgement
We thank Marmara University (Division of Medical Oncology and Department of Medical Genetics) and Istanbul Aydin University (Health Sciences Research Laboratory) for providing the case- control samples.
Funding
This work has been supported by Marmara University Scientific Research Projects Coordination Unit under grant number SAG-C- DRP-120619-0222.
Ethical approval
All human participants involving procedures performed were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by the Ethics Committee of Medical Faculty of Marmara University (Ethical Approval No: 09.2019.204).
References
- Bray F, Ferlay J, Soerjomataram I, et Global cancer statistics 2018 GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a cancer journal for clinicians. 2018; 68: 394-424.
- Harbeck N, Penault-Llorca F, Cortes J, et Breast cancer. Nat Rev Dis Primers. 2019; 5: 66.
- Al-Mahmood S, Sapiezynski J, Garbuzenko OB, et al. Metastatic and triple-negative breast cancer challenges and treatment options. Drug Deliv Transl Res. 2018; 8: 1483-1507.
- Dimitrakopoulos FD, Antonacopoulou AG, Kottorou AE, et Expression of Intracellular Components of the NF- κB Alternative Pathway NFKB2, RelB NIK and BCL3 is Associated with Clinical Outcome of NSCLC Patients. Sci Rep. 2019; 9: 14299.
- Maldonado V, Melendez-Zajgla J. Role of BCL-3 in solid tumors. Mol Cancer. 2011; 10: 152.
- Kim HJ, Hawke N, Baldwin AS. NF-kappaB and IKK as therapeutic targets in Cell Death Differ. 2006; 13: 738- 747.
- Legge DN, Chambers AC, Parker CT, et The role of B-Cell Lymphoma-3 BCL-3 in enabling the hallmarks of cancer implications for the treatment of colorectal carcinogenesis. Carcinogenesis. 2020; 41: 249-256.
- Puvvada SD, Funkhouser WK, Greene K, et al. NF-kB and BCL-3 activation are prognostic in metastatic colorectal Oncology. 2010; 78: 181-188.
- Wakefield A, Soukupova J, Montagne A, et BCL3 selectively promotes metastasis of ERBB2-driven mammary tumors. Cancer Res. 2013; 73: 745-755.
- Chen X, Cao X, Sun X, et BCL-3 regulates TGFβ signaling by stabilizing Smad3 during breast cancer pulmonary metastasis. Cell Death Dis. 2016; 7: e2508.
- Tu K, Liu Z, Yao B, et BCL-3 promotes the tumor growth of hepatocellular carcinoma by regulating cell proliferation and the cell cycle through cyclin D1. Oncol Rep. 2016; 35: 2382-2390.
- Hanson C, Cairns J, Wang L, et al. Computational discovery of transcription factors associated with drug response. Pharmacogenomics J. 2016; 16: 573-582.
- World Medical Association. World Medical Association Declaration of Helsinki ethical principles for medical research involving human subjects. JAMA. 2013; 310: 2191-2194.
- Verhoeft KR, Ngan HL, Lui The cylindromatosis CYLD gene, head, and neck tumorigenesis. Cancers Head Neck. 2016; 1: 10.
- Cogswell PC, Guttridge DC, Funkhouser WK, et Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for BCL-3. Oncogene. 2000; 19: 1123-1231.
- Westerheide SD, Mayo MW, Anest V, et al. The putative oncoprotein BCL-3 induces cyclin D1 to stimulate G1 Mol Cell Biol. 2001; 8428-8436.
- Pratt MA, Bishop TE, White D, et al. Estrogen withdrawal- induced NF-kappaB activity and BCL-3 expression in breast cancer cells roles in growth and hormone independence. Mol Cell Biol. 2003; 23: 6887-6900.
- Kashatus D, Cogswell P, Baldwin AS. Expression of the BCL-3 proto-oncogene suppresses p53 Genes Dev. 2006; 20: 225-235.
- Palmer S, Chen YH. BCL-3 a multifaceted modulator of NF- kappaB-mediated gene Immunol Res. 2008; 42: 210-218.
- Choi HJ, Lee JM, Kim H, et BCL3-dependent stabilization of CtBP1 is crucial for the inhibition of apoptosis and tumor progression in breast cancer. Biochem Biophys Res Commun. 2010; 400: 396-402.
- Urban BC, Collard TJ, Eagle CJ, et al. BCL-3 expression promotes colorectal tumorigenesis through activation of AKT signalling. Gut. 2016; 65: 1151-1164.
- Wang W, Nag SA, Zhang R. Targeting the NFκB signaling pathways for breast cancer prevention and Curr Med Chem. 2015; 22: 264-289.
- Saamarthy K, Björner S, Johansson M, et Early diagnostic value of BCL-3 localization in colorectal cancer. BMC Cancer. 2015; 15: 341.
- Huo J, Chen X, Zhang H, et BCL-3 promotes proliferation and chemosensitivity in BL1 subtype of TNBC cells. Acta Biochim Biophys Sin Shanghai. 2018; 50: 1141-1149.
- Hu L, Bai Z, Ma X, et The Influence of BCL-3 Expression on Cell Migration and Chemosensitivity of Gastric Cancer Cells via Regulating Hypoxia-Induced Protective Autophagy. J Gastric Cancer. 2020; 20: 95-105.
- Massoumi R, Chmielarska K, Hennecke K, et Cyld inhibits tumor cell proliferation by blocking BCL-3-dependent NF- kappaB signaling. Cell. 2006; 125: 665-677.
- Rivlin N, Koifman G, Rotter V. p53 orchestrates between normal differentiation and cancer. Semin Cancer Biol. 2015; 32: 10-17.
- Mrozik KM, Blaschuk OW, Cheong CM, et al. N-cadherin in cancer metastasis its emerging role in haematological malignancies and potential as a therapeutic target in cancer. BMC Cancer. 2018; 18: 939.
- Lin Z, Hegarty JP, Yu W, et al. Identification of disease- associated DNA methylation in B cells from Crohn's disease and ulcerative colitis patients. Dig Dis Sci. 2012; 57: 3145-
- Wang SM, Ye M, Ni SM, et al. Effect of COX-2 inhibitor on the expression of BCL-3 and cyclin D1 in human colon cancer cell line SW480. Chinese. 2010; 13: 612-615.