Hepatitis B, D, C, and HCC viruses in Tajikistan
Author'(s): Abdusamad Dustov* and Zebunisso Khakimova
Institute of Gastroenterology, Ministry of Health of the Republic of Tajikistan.
*Correspondence:
Abdusamad Dustov, Institute of Gastroenterology, Ministry of Health of the Republic of Tajikistan, E-mail: dustovsamad@gmail.com.
Received: 02 January 2018 Accepted: 14 February 2018
Citation: Abdusamad Dustov, Zebunisso Khakimova. Hepatitis B, D, C, and HCC viruses in Tajikistan. Gastroint Hepatol Dig Dis. 2018; 1(1) 002: 1-4.
Keywords
Introduction
Hepatitis B virus (HBV) and hepatitis C virus (HCV) are common etiological agents of viral hepatitis. Approximately 350 million people are infected with HBV worldwide and the World Health Organization (WHO) estimates that approximately 170 million people are infected with HCV. Chronic HBV and/or HCV infection can progress to liver cirrhosis and hepatocellular carcinoma (HCC). HCV displays a high degree of genetic variability and is classified into six major genotypes that show sequence similarities of only 66-69% [1]. There has been no report on the prevalence of the hepatitis B and C viruses in Tajikistan. Therefore, the aim of the present study was to investigate prevalence viral hepatitis D and C among population and the genotypic prevalence and clinical significance of HCV, HBV, and/ or HDV among chronic hepatitis patients with and without liver cirrhosis and/or HCC in Tajikistan. Furthermore, the association between coinfection patterns or HBV specific mutations and disease progression were also analyzed in this study.
Methods
Patients Sera were obtained from 124 consecutive chronic liver diseases patients over a period of 6 months between February to August 2008. Patients in this study were classified into two clinical groups (i) Chronic hepatitis: patients with constantly high serum alanine aminotransferase level (ALT), and (ii) Liver cirrhosis and HCC: patients, as diagnosed clinically based on ultrasonography, detection of serological tumor markers (AFP) and in a few cases histologically.
Serological Testing
Anti-HCV, HBsAg, HBeAg, anti-HBc and anti-HBs were determined by chemiluminescence with commercial assay kits (Fujirebio, Inc., Tokyo, Japan). HCV core antigen (HCVcAg), was measured [2], using enzyme immunoassay (Fujirebio, Inc.). Delta antibody (anti-HDV) was assessed only in HBsAg positive specimens using the recombinant HDVAg [3] diagnostic kit ELISA Anti-HDV (RPC Diagnostics Systems, Nizhnyi Novgorod, Russia). Biochemical markers, that is, ALT, AST, Alkaline Phosphatase, cholestrol, bilurubin were measured in all samples at the local hospital. Genotyping of HCV Total RNA was extracted from serum, reverse transcribed into cDNA using random hexamer primer as described previously [4]. Confirmation of HCV RNA in the samples was carried by amplifying highly conserved 50UTR region and HCV genotypes were determined for both; structural (E1/Core) and nonstructural (NS5B) viral genes using either or both genotyping PCR [4,5] and/or direct sequencing with genotype-universal primers [6]. Screening for HCV Type RF1_2k/1b screening for natural recombinant type RF1_2k/1b strain was performed by the newly developed sensitive and specific method allowing detection up to 10 copies/assay (100 copies/ml) of the target sequence even if it is present in minority in mixed heterogeneous template. Briefly, the primers were designed to amplify a part of NS2 coding region between nt. 2986 and 3270 spanning the intergenotypic breakpoint described for RF1_2k/1b [7]. Target product’s size was 347 base pair (bp) in the first round of PCR and 300 bp in the second round of the ‘‘hemi-nested’’ PCR.
Genotyping of HBV and HDV
All of the HBsAg-positive samples were subjected to genotyping by commercial EIA kit (Institute of Immunology Co., Ltd, Tokyo, Japan). The method allows discrimination among the seven major HBV genotypes (A-G) by monoclonal antibodies targeted to the preS2 epitopes [8]. HBV DNA was extracted by QIAamp DNA Blood Mini Kit (Qiagen, Inc., Hilden, Germany) from 100 ml of all HBsAg-positive sera. Complete genome was successfully sequenced for 7 strains and a part of the HBV core and short S region was sequenced for other 30 strains using previously reported primers [9]. HDV RNA was extracted from anti-HDV positive samples and transcribed reversely into a cDNA using random primer as previously described for HCV [4]. A part of HDVAg coding region of the HDV was amplified using specific primers described previously [10] and sequenced directly. Phylogenetic Analysis PCR products were sequenced directly with the Prism Big Dye (Applied Biosystems, Foster City, CA) in an ABI 3100 DNA automated sequencer. All sequences were analyzed in both forward and reverse directions.Complete and partial HBV genomes were assembled using GENETYX version 11.0 (GENETYX, Tokyo, Japan). The sequences for phylogenetic analysis were retrieved from DDBJ/EMBL/GeneBank corresponding to the accession numbers mentioned in the trees. Alignment was accomplished using CLUSTAL W and NJ tree were constructed with Tamura- Nei distance correction model using online tools of the HCV database [11].
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession number(s) AB330313 to AB330383.
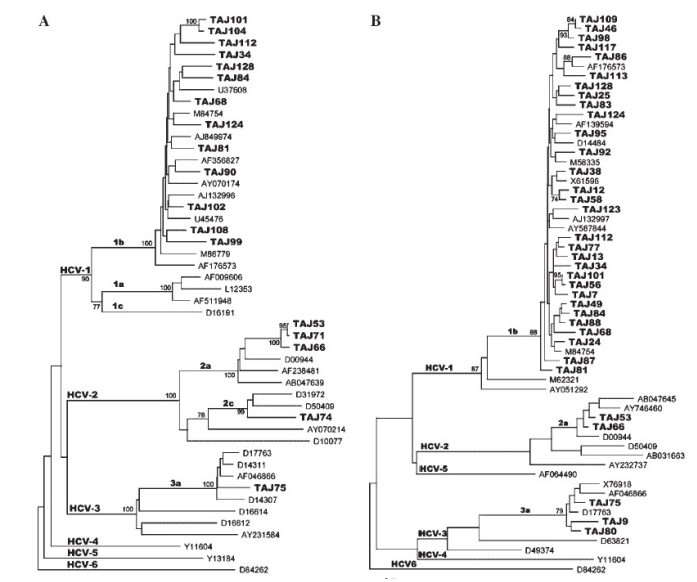
Figure 1A: Phylogenetic analysis of the hepatitis C virus (HCV) partial E1 region, sequences from Tajikistan and other countries. Tajikistan isolates (e.g., TAJ74, TAJ75, etc.) were subject to bootstrap resampling with all available sequences in E1 region retrieved from the EMBL/ DDBJ/Gene Bank database. Closest neighbors used for phylogenetic tree are indicated under the corresponding accession numbers from DDBJ/ EMBl/Gene Bank. B: Phylogenetic analysis of the HCV partial NS5B region, sequences from Tajikistan and other countries. Tajikistan isolates (e.g., TAJ108, TAJ75, etc.) were subject to bootstrap resampling with all available sequences in NS5B region retrieved from the EMBL/DDBJ/ Gene Bank database. Closest neighbors used for phylogenetic tree are indicated under the corresponding accession numbers from DDBJ/EMBl/ Gene Bank.
Stastical Analysis
Statistical differences were evaluated by Fisher’s exact probability test and Chi-square test with Yates’ correction, where appropriate, using the STATA software version 8.0 (Stata Corp. LP, College Station, TX). Differences were considered significant for P values less than 0.05.
Results
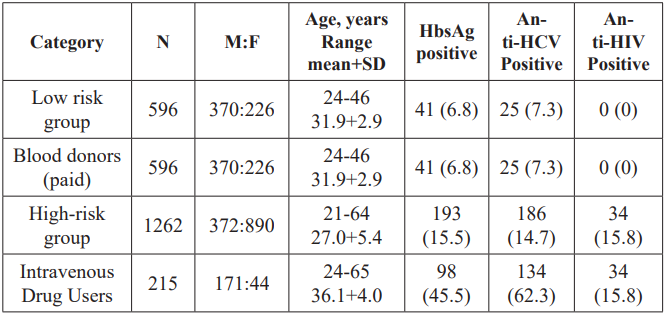
Table 1: Seroprevalence of HbsAG, anti HCV and anti-HIV among various groups in Tajikistan.
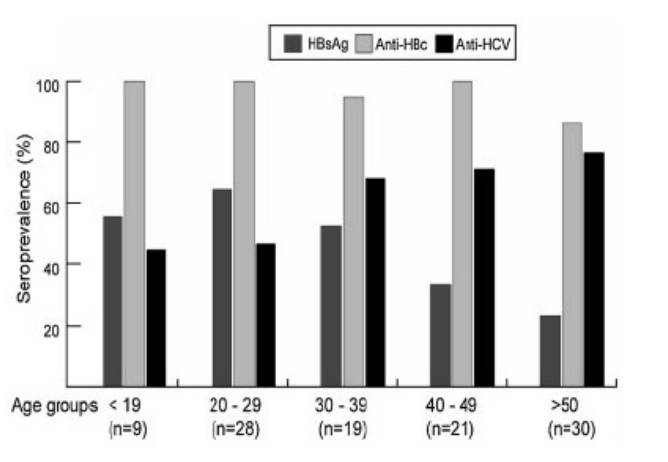
Figure 2: Comparative prevalence of viral hepatitis markers in different age groups of studied chronic liver disease patients.
The HCV, HBV, and HDV Prevalence and the Co-Infection Patterns among 124 hepatitis patients, 84 (67.7%) were assigned into chronic hepatitis (group 1) and 40 (32.2%) into liver cirrhosis/ HCC (group 2). The overall male to female ratio was 1.4, and the mean age (standard deviation) was 38.4 (14.9). These estimations are summarized in Table 2.
There was no significant difference in age between both groups. The ALT, AST, and total bilirubin, were high without significant difference between the both groups. Overall alkaline phosphatases (ALP) levels were within normal range in both groups. HCV- infection was defined upon detection of HCV RNA or HCVcAg positive and HCV recovered as anti-HCV positive but negative for HCVcAg or HCV RNA. HCV core antigen (HCVcAg) was measured in serum, where the cutoff value was set tentatively at
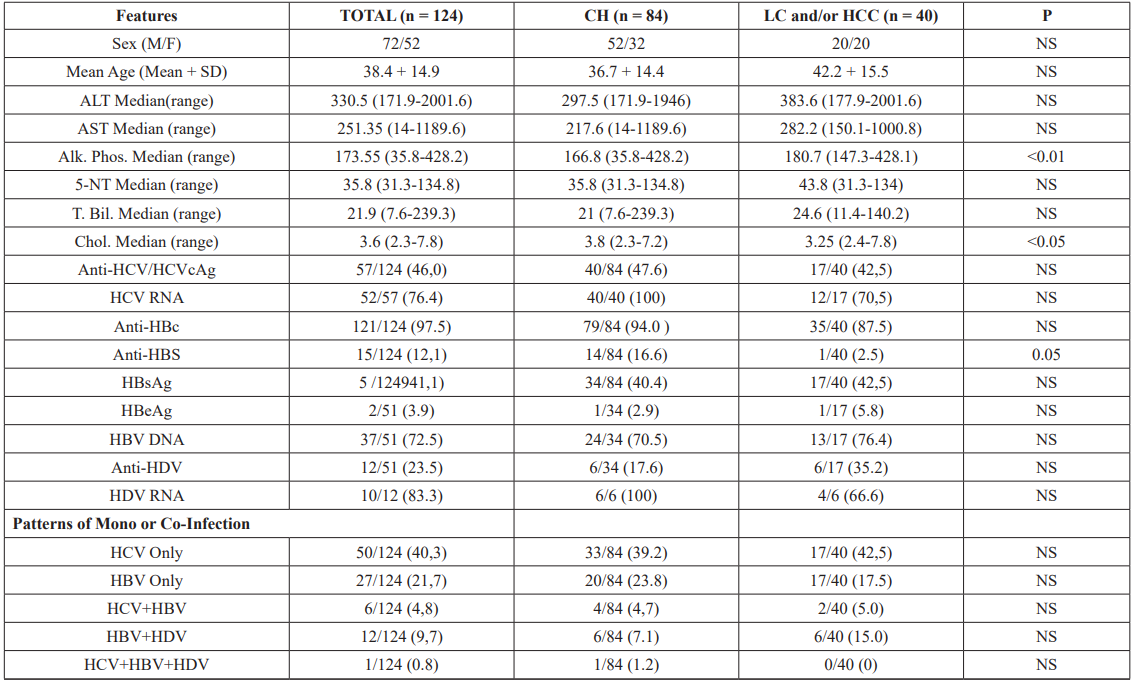
Table 2: Clinical Feature and Hepatitis Markers among 124 Subjects With Chronic Liver Disease in Tajikistan.
(Normal ranges: ALT: 33.3-200.1 nM/l., AST: 27.8-150.1 nM/l., ALP: 44.5-219.6 nM/l., 5’NT: 8.95-62.7 nM/l., T.Bilu: 8.5-20.5 mM/l., Chol 3-6.2 nM/l).
50 fmol/L. In this study 15 out of 68 anti-HCV positive samples were negative for HCV RNA. The HCVcAg was positive in only 4 of the 15 cases, suggesting that HCV RNA was degraded in these samples, while remaining 11 cases had resolved acute HCV infection. HCV infection was found to be high in both groups (group 1.47.6%and group 2.42.5%). HCV RNA was detected in 91% of cases (100% in group 1 and 70.5% in group 2). Anti-HBc was found to be high in both groups (94% and 87.5%, respectively). Although seroprevalence of anti-HBs was very low (12.1%) in the studied population but significantly high in group 1 (16.6%) than group 2 (P.0.0356) while a few cases were positive for anti-HBeAg (2.9% and 5.8%, respectively). Positivity for HBsAg was regarded as indicating HBV infection.
The seroprevalence of HBsAg was equally high in both groups, that is, group 1.40.4% and group 2.42.5%, whereas DNA positivity was 70.5% and 76.4%, respectively. Overall anti-HDV seroprevalence was 23.5%, 12 out of 51 HBsAg positive cases, relatively higher in group 2 (35.2%) compared to group 1 (17.6%) P.0.1990.HDV viremia was detected in 83.3% of cases (100% in group 1 and 66.6% in group 2) (Table 1). Based on the serological findings for hepatitis viruses, the patients were categorized into five groups, HCV only (40.3%), HBV only (21.7%), HCV/HBV (4.8%), HBV/ HDV (9.7%), and HCV/HBV/HDV (0.8%) in Tajikistan (Table 1).

Figure 3: Phylogenetic analysis of the hepatitis B virus (HBV) eight full genomes, sequences from Tajikistan and other countries. Tajikistan isolates (e.g., TAJ5, TAJ6, etc.) were subject to bootstrap resampling with all available full genome sequences retrieved from the EMBL/DDBJ/ Gene Bank database. Closest neighbors used for phylogenetic tree are indicated under the corresponding accession numbers from DDBJ/ EMBl/ Gene Bank.
When the seropositivity of hepatitis markers was analyzed in relation to age (Figure 1), it was found that the prevalence of HBsAg was relatively high in younger group (<29 years old) and tended to decline with age. The opposite trend was observed in anti-HCV prevalence, which was increasing with the age of the patients. The seroprevalence of anti-HBc was high in all age groups, indicating that most patients had past HBV infection during childhood.
HBV, HDV and HCV Genotypes
HCV genotyping was carried by two independent methods, that is, by type specific primers for core, NS5B region and phylogenetic analyses based on nucleotide sequences in E1 or NS5B region. No discrepancy was observed among the results between either method (Figure 2A,B), however, one case (TAJ74) was determined as HCV genotype 2c (HCV/2c) only by the phylogenetic analysis in E1 region (Figure 2A). A total of 7.6% of cases were untypeable by one of either method (Table 2). Overall, HCV/1bwas a predominant genotype (84.6%) in Tajikistan, followed by HCV/3a (7.6%), 2a (5.7%), and 2c (1.9%). None of the HCV- infected in this study carried recombinant 2k/1b strain. Other potential forms of recombination were also excluded by matching result of genotyping based on both structural and nonstructural genomic parts (Table 2). Interestingly, all 12 LC/HCC patients had HCV/1b, not 3a or 2a (Table 3). HBV genotype D (HBV/D) was the predominant genotype (94.1%) in both groups, that is, group 1=97% and group 2=88.2%, followed by genotype A (2.9% and 11.7%, respectively). HBV genotypes were determined in 45 (88%) of 51 HBsAg-positive cases by EIA. The entire HBV genome was sequenced successfully for seven cases (Figure 3), and the precore/core promoter region was sequenced in the other 25 cases. The remaining five cases were only positive by PCR for a short S region. The full genome analysis revealed that of the four HBV/D strains, two belonged to subgenotype D1, and the remaining two to subgenotype D2. All three HBV/A strains in this study were belonged to subgenotype Ae.
Variability of HBV in Enhance II, Basal Core Promoter (BCP), Pre-C/C Region
The prevalence of the nucleotide substitutions along the 33 sequence bearing the enhancer II/core promoter/ precore/core regions of the HBV genome are summarized for both clinical groups (22 group 1 vs. 11 group 2) in the Table 4. Due to the small numbers of patients with liver cirrhosis (n.8) and HCC (n.3), these were combined in the ‘‘Group 2’’ to enable the comparative analysis with chronic hepatitis patients without the complications; ‘‘Group 1.’’ The frequency of A1757 was high in both of the groups without the difference significant statistically. The T1762/A1764 double mutation was more frequent in the Group 2 and this was significant; P.0.0004. The difference in prevalence of the A1896, A1899, and T1912 mutations did not reach statistical significance between the groups.
Discussion
In the present study, the seroprevalence and molecular-enetic characteristics of the hepatitis viruses were investigated among chronic hepatitis (Group 1) and liver cirrhosis/HCC (Group 2) patients in Tajikistan. The HCV seroprevalence was equally high in both of the groups (47.6% and 42.5%, respectively) followed by HBsAg (40.4% and 42.5%, respectively). These results are slightly higher than the previous studies carried in neighboring Uzbekistan, where anti-HCV was detected in 26.8-41.9% and HBsAg in 25.6% of chronic hepatitis patients [12,13], such a difference may be explained by the different ages of studied patients different collecting time and also regional characteristics. A rather old study carried on the healthy population in Tajikistan and Azerbaijan showed that HBsAg detection was significantly higher in Tajikistan (7.2%) than in Azerbaijan (2.8%) suggesting that HBV is endemic in this country [14]. Tajikistan introduced universal immunization against HBV for newborn children since 2002. Although, new anti-viral drugs have the potential to alter the natural progression of disease but their high cost prevents the mass use of therapy in Tajikistan. One of the interesting observations in this study was the trend of seroprevalence of anti-HCV to increase and HBsAg do decrease with the age of the patients. Interestingly, the prevalence of anti-HBc was very high regardless the age of the patients, indicating that most of the patients had present or past infection of HBV.
The age-related decline of the HBsAg prevalence observed in this study may be explained by either; seroclearance of HBsAg due to natural seroconversion [15] or by death of the HBV-infected patients at age younger than those with HCV, due to earlier development of the end stage liver disease [16-18]. Alternatively, suppressive effect of the concurrent HCV infection might have caused the loss of the HBV antigenia [19-21]. HCV/1b has been found as the dominant genotype (84.6%) in Tajikistan, followed by 3a (7.6%), 2a (5.7%), and 2c (1.9%). The similar epidemiological situation was also described in the former Soviet Union Republics (Russia, Belarus, Moldova, and Uzbekistan) [13,22,23] as well as in most of Asian countries, namely Mongolia [24], Taiwan [25], China [26], and Japan [27] where genotype 1b was predominant. The HCV core antigen (HCVcAg) has significant correlation with HCVRNA [28,29]. In this study 15 out of the 68 anti-HCV positive simples, had the HCV RNA undetectable by PCR method, however only 4 of the 15 cases had the HCVcAg detectable, suggesting that the viral RNA has degraded in these samples prior study [30], while the remaining 11 cases may have had an acute HCV infection resolved in the past. HBV genotypes have a characterstic geographical distribution [31,32]. Particularly, genotype D is widespread, predominating in the Mediterranean area and in the Middle East region. Genotype A is present in Europe, India, Africa, and North America. The present results have shown that 94% of HBV isolates in Tajikistan belonged to genotype D and 5.8% to genotype A. Similar epidemiological situation was observed in a previous study carried in Uzbekistan, where HBV/D found in 87% and HBV/A in 13% of HBV infected patients [33]. Complete genome analysis of the four HBV/D strains determined in present study allowed distinction into two subgenotypes; D1 and D2. Previous reports indicate that HBV/D subgenotypes have no specific geographical distribution; however strains from the Middle East mainly belong to subgenotype D1 and those from India, Russia and Europe to D2 [32]. No specific clustering was observed in Tajikistan strains, which is in agreement with previous observations regarding HBV/D molecular epidemiology. HBV genotype A has three major geographical clusters, the European-North American (Ae), the Afro-Asian (Aa) [34], and the Central African (Ac) [35]. The HBV genotype A strains isolated in this study were all belonged to the subgenotype Ae. Increasing evidence suggests that specific HBV genomic mutations may increase the risk of HCC in the patients infected chronically. Particularly, a high incidence of the T1762/A1764 double mutation was found among HBV/B or C infected Asian population [36] and with HBV/E infected African patients [37]. Similarly, in the present study, the frequency of the double mutation was significantly high in group 2 (6HBV/D and 2 HBV/A) as compared to group 1 (1HBV/D and 1HBV/A). In vitro studies [18,38], indicated that the T1762/A1764 mutants show enhanced replication and might be associated with high incidence of HCC. The prevalence of coinfection with HBV and HDV found to be relatively higher in group 2 compared to group 1 (15% vs. 7.1%), however the difference was not statistically significant possibly due to the small size of cases (P.0.1990). Previous studies have demonstrated that the coinfection with these viruses was also contributing in high HCC incidence in Mongolia [16]. It is concluded that HBV and HCV are the major cause of viral hepatitis in Tajikistan. Among HBV-infected patients, the T1762/A1764 mutations were associated with liver cirrhosis/HCC in this population. We have sent 60 blood samples from patients with hepatitis B and D to the CDC (Centers for Disease Control and Prevention) Atlanta, GA, USA. We have received data on the prevalence of the HBV and HDV genotype in Tajikistan, the work on our samples continues. Molecular genetic study of the HDV in Tajikistan has shown that HDV genotype 1 occurs in 96% (58/ 60 examined). According to this literature, HDV genotype 1 is found in Asia and in only 25% of these patients the antiviral therapy was successful [39].
References
- Pawlotsky JM. A. Hepatitis C virus genetic variability: Pathogenic and clinical implications. Clin Liver Dis. 2003; 7: 45-66.
- Aoyagi K, Ohue C, Iida K, et Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999; 37: 1802-1808.
- Semiletov Iu A, Pimenov VK, Ianina MV, et al. Test for specific antibodies to virus hepatitis delta based on synthetic peptides. Vopr Virusol. 2002; 47: 21-24.
- Ohno O, Mizokami M, Wu RR, et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and J Clin Microbiol. 1997; 35: 201-207.
- Hashimoto M, Chayama K, Tubota A, et Typing six major hepatitis C virus genotypes by polymerase chain reaction using primers derived from nucleotide sequences of the NS5 region. Int Hepatol Commun. 1996; 4: 263-267.
- Tanaka Y, HanadaK, Mizokami M, et al. Inaugural article: A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc Natl Acad Sci USA. 2002; 99: 15584-15589.
- Kalinina O, Norder H, Mukomolov S, et al. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol. 2002; 76: 4034-4043.
- Usuda S, Okamoto H, Iwanari H, et al. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J Virol Methods. 1999; 80: 97-112.
- Sugauchi F, Mizokami M, Orito E, et al. A novel variant genotypeCofhepatitisBvirus identified in isolates from Australian Aborigines: Complete genome sequence and phylogenetic relatedness. J Gen Virol. 2001; 82: 883-892.
- Nakano T, Shapiro CN, Hadler SC, et Characterization of hepatitisDvirus genotype III among Yucpa Indians in Venezuela. J Gen Virol. 2001; 82: 2183-2189.
- Kuiken C, Mizokami M, Deleage G, et al. Hepatitis C databases, principles and utility to researchers. Hepatology. 2006; 43: 1157-1165.
- Ruzibakiev R, Kato H, Ueda R, et al. Risk factors and seroprevalence of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus infection in uzbekistan. Intervirology. 2001; 44: 327-332.
- Kurbanov F, Tanaka Y, Sugauchi F, et al. 2003. Hepatitis C virus molecular epidemiology in Uzbekistan. J Med Virol. 2003; 69: 367-375.
- Vorozhbieva TE, Iasinskii AV, Alieva G, et Characteristics of the distribution of the markers of hepatitis B virus infection among the healthy population of the Tadzhik SSR and Azerbaijan SSR. Zh Mikrobiol Epidemiol Immunobiol. 1985; 10: 35-39.
- HepatitisBvirus infection.NEnglJMed. 1997; 337: 1733-1745.
- Oyunsuren T, Kurbanov F, Tanaka Y, et al. 2006. High frequency of hepatocellular carcinoma inMongolia; association with mono-, or co-infection with hepatitis C, B, and delta viruses. J Med Virol. 2006; 78: 1688-1695.
- Kurbanov F, Tanaka Y, Elkady A, et Tracing hepatitis Cand Delta viruses to estimate their contribution in HCC rates in Mongolia. J Viral Hepat. 2007; 14: 667-674.
- Idrees M, Ur Rehman I, Manzoor S, et Evaluation of three different hepatitis C virus typing methods for detection of mixed- genotype infections. J Dig Dis. 2011; 12: 199-203.
- Sheen IS, Liaw YF, LinDY, et Role of hepatitisCand delta viruses in the termination of chronic hepatitis B surface antigen carrier state: A multivariate analysis in a longitudinal follow-up study. J Infect Dis. 1994; 170: 358-361.
- Chu CJ, Hussain M, Lok Quantitative serum HBV DNA levels during different stages of chronic hepatitis B infection. Hepatology.2002; 36: 1408-1415.
- Quarleri Core promoter: a critical region where the hepatitis B virus makes decisions. World J Gastroenterol. 2014; 20: 425-435.
- Viazov S, Kuzin S, Paladi N, et Hepatitis C virus genotypes in different regions of the former Soviet Union (Russia, Belarus, Moldova, and Uzbekistan). J Med Virol. 1997; 53: 36-40.
- Shustov AV, Kochneva GV, Sivolobova GF, et al. 2005. Molecular epidemiology of the hepatitis C virus inWestern Siberia. J Med Virol. 2005; 77: 382-389.
- Lvov DK, Samokhvalov EI, Tsuda F, et al. Prevalence of hepatitis virus and distribution of its genotypes in Northern Eurasia. Arch Virol. 1996; 141: 1613-1622.
- Yu ML, Chuang WL, Chen SC, et al. Changing prevalence of hepatitis C virus genotypes: Molecular epidemiology and clinical implications in the hepatitis C virus hyperendemic areas and a tertiary referral center in Taiwan. J Med Virol. 2001; 65: 58-65.
- Suzuki K, Mizokami M, Cao K, et al. Prevalence of hepatitis C virus infection in Nanjing, southern Eur J Epidemiol. 1997; 13: 511-515.
- Noguchi S, Sata M, Suzuki H, et Routes of transmission of hepatitis C virus in an endemic rural area of Japan.Molecular epidemiologic study of hepatitis Cvirus infection. Scand J Infect Dis. 1997; 29: 23-28.
- Tanaka E, Ohue C, Aoyagi K, et al. Evaluation of a new enzyme immunoassay for hepatitis C virus (HCV) core antigen with clinical sensitivity approximating that of genomic amplification of HCV Hepatology. 2000; 32: 388-393.
- Chen GF, Wei L, Chen J, et al. Will Sofosbuvir/Ledipasvir (Harvoni) Be Cost-Effective and Affordable for Chinese Patients Infected with Hepatitis C Virus? An Economic Analysis Using Real-World Data. PLoS One. 2016; 11:
- Tanaka Y, Takagi K, Fujihara T, et High stability of enzyme immunoassay for hepatitis C virus core antigen-evaluation before and after incubation at room temperature. Hepatol Res. 2003; 26: 261-267.
- Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003; 46: 329-338.
- Norder H, Courouce AM, Coursaget P, et Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004; 47: 289-309.
- Kato H, Ruzibakiev R, Yuldasheva N, et Hepatitis B virus genotypes in Uzbekistan and validity of two different systems for genotyping. J Med Virol. 2002; 67: 477-483.
- Sugauchi F, Kumada H, Acharya SA, et al. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype J Gen Virol. 2004; 85: 811- 820.
- Kurbanov F, Tanaka Y, Fujiwara K, et al. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in J Gen Virol. 2005; 86: 2047-2056.
- Kao JH, Chen PJ, Lai MY, et Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003; 124: 327-334.
- Baptista M, Kramvis A, Kew High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology. 1999; 29: 946-953.
- Buckwold VE, Xu Z, Chen M, et 1996. Effects of a naturally occurring mutation in the hepatitis B virus basal corepromoter on precore gene expression and viral replication. J Virol. 1996; 70: 5845-5851.
- Bogomolov P, Alexandrov A, Voronkova N, et al. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: First results of a phase Ib/IIa J Hepatol. 2016; 65: 490- 498.